Large-scale identification of protein-protein interaction of Escherichia coli K-12
- PMID: 16606699
- PMCID: PMC1457052
- DOI: 10.1101/gr.4527806
Large-scale identification of protein-protein interaction of Escherichia coli K-12
Abstract
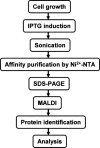
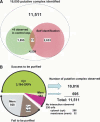
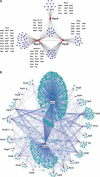
Protein-protein interactions play key roles in protein function and the structural organization of a cell. A thorough description of these interactions should facilitate elucidation of cellular activities, targeted-drug design, and whole cell engineering. A large-scale comprehensive pull-down assay was performed using a His-tagged Escherichia coli ORF clone library. Of 4339 bait proteins tested, partners were found for 2667, including 779 of unknown function. Proteins copurifying with hexahistidine-tagged baits on a Ni2+-NTA column were identified by MALDI-TOF MS (matrix-assisted laser desorption ionization time of flight mass spectrometry). An extended analysis of these interacting networks by bioinformatics and experimentation should provide new insights and novel strategies for E. coli systems biology.
Figures

References
-
- Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H., Datsenko K.A., Tomita M., Wanner B.L., Mori H., Tomita M., Wanner B.L., Mori H., Wanner B.L., Mori H., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knock-out mutants—the Keio collection. Mol. Syst. Biol. 2006 http://www.nature.com/msb/journal/v2/n1/full/msb4100050.html - PMC - PubMed
-
- Bader G.D., Hogue C.W., Hogue C.W. Analyzing yeast protein–protein interaction data obtained from different sources. Nat. Biotechnol. 2002;20:991–997. - PubMed
-
- Barabasi A.L., Albert R., Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. - PubMed
-
- Brandi A., Pon C.L., Gualerzi C.O., Pon C.L., Gualerzi C.O., Gualerzi C.O. Interaction of the main cold shock protein CS7.4 (CspA) of Escherichia coli with the promoter region of hns. Biochimie. 1994;76:1090–1098. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
