Artificial control of gene expression in mammalian cells by modulating RNA interference through aptamer-small molecule interaction
- PMID: 16606868
- PMCID: PMC1440907
- DOI: 10.1261/rna.2299306
Artificial control of gene expression in mammalian cells by modulating RNA interference through aptamer-small molecule interaction
Abstract
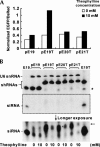
Recent studies have uncovered extensive presence and functions of small noncoding RNAs in gene regulation in eukaryotes. In particular, RNA interference (RNAi) has been the subject of significant investigations for its unique role in post-transcriptional gene regulation and utility as a tool for artificial gene knockdown. Here, we describe a novel strategy for post-transcriptional gene regulation in mammalian cells in which RNAi is specifically modulated through RNA aptamer-small molecule interaction. Incorporation of an RNA aptamer for theophylline in the loop region of a short hairpin RNA (shRNA) designed to silence fluorescent reporter genes led to dose-dependent inhibition of RNAi by theophylline. shRNA cleavage experiments using recombinant Dicer demonstrated that theophylline inhibited cleavage of an aptamer-fused shRNA by Dicer in vitro. Inhibition of siRNA production by theophylline was also observed in vivo. The results presented here provide the first evidence of specific RNA-small molecule interaction affecting RNAi, and a novel strategy to regulate mammalian gene expression by small molecules without engineered proteins.
Figures



References
-
- Bayer T.S., Smolke C.D. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat. Biotechnol. 2005;23:337–343. - PubMed
-
- Bogenhagen D.F., Brown D.D. Nucleotide sequences in Xenopus 5S DNA required for transcription termination. Cell. 1981;24:261–270. - PubMed
-
- Brummelkamp T.R., Bernards R., Agami R. A system for stable expression of short interfering RNAs in mammalian cells. Science. 2002;296:550–553. - PubMed
-
- Buskirk A.R., Landrigan A., Liu D.R. Engineering a ligand-dependent RNA transcriptional activator. Chem. Biol. 2004;11:1157–1163. - PubMed
-
- Chiu Y.L., Rana T.M. RNAi in human cells. Basic structural and functional features of small interfering RNA. Mol. Cell. 2002;10:549–561. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
