A phylogenomic gene cluster resource: the Phylogenetically Inferred Groups (PhIGs) database
- PMID: 16608522
- PMCID: PMC1523372
- DOI: 10.1186/1471-2105-7-201
A phylogenomic gene cluster resource: the Phylogenetically Inferred Groups (PhIGs) database
Abstract
Background: We present here the PhIGs database, a phylogenomic resource for sequenced genomes. Although many methods exist for clustering gene families, very few attempt to create truly orthologous clusters sharing descent from a single ancestral gene across a range of evolutionary depths. Although these non-phylogenetic gene family clusters have been used broadly for gene annotation, errors are known to be introduced by the artifactual association of slowly evolving paralogs and lack of annotation for those more rapidly evolving. A full phylogenetic framework is necessary for accurate inference of function and for many studies that address pattern and mechanism of the evolution of the genome. The automated generation of evolutionary gene clusters, creation of gene trees, determination of orthology and paralogy relationships, and the correlation of this information with gene annotations, expression information, and genomic context is an important resource to the scientific community.
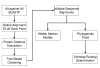
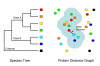
Discussion: The PhIGs database currently contains 23 completely sequenced genomes of fungi and metazoans, containing 409,653 genes that have been grouped into 42,645 gene clusters. Each gene cluster is built such that the gene sequence distances are consistent with the known organismal relationships and in so doing, maximizing the likelihood for the clusters to represent truly orthologous genes. The PhIGs website contains tools that allow the study of genes within their phylogenetic framework through keyword searches on annotations, such as GO and InterPro assignments, and sequence similarity searches by BLAST and HMM. In addition to displaying the evolutionary relationships of the genes in each cluster, the website also allows users to view the relative physical positions of homologous genes in specified sets of genomes.
Summary: Accurate analyses of genes and genomes can only be done within their full phylogenetic context. The PhIGs database and corresponding website http://phigs.org address this problem for the scientific community. Our goal is to expand the content as more genomes are sequenced and use this framework to incorporate more analyses.
Figures


References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Research Materials

