Mutations in the endodomain of Sindbis virus glycoprotein E2 define sequences critical for virus assembly
- PMID: 16611906
- PMCID: PMC1472013
- DOI: 10.1128/JVI.80.9.4458-4468.2006
Mutations in the endodomain of Sindbis virus glycoprotein E2 define sequences critical for virus assembly
Abstract
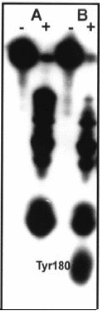
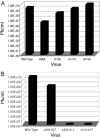
Envelopment of Sindbis virus at the plasma membrane is a multistep process in which an initial step is the association of the E2 protein via a cytoplasmic endodomain with the preassembled nucleocapsid. Sindbis virus is vectored in nature by blood-sucking insects and grows efficiently in a number of avian and mammalian vertebrate hosts. The assembly of Sindbis virus, therefore, must occur in two very different host cell environments. Mammalian cells contain cholesterol which insect membranes lack. This difference in membrane composition may be critical in determining what requirements are placed on the E2 tail for virus assembly. To examine the interaction between the E2 tail and the nucleocapsid in Sindbis virus, we have produced substitutions and deletions in a region of the E2 tail (E2 amino acids 408 to 415) that is initially integrated into the endoplasmic reticulum. This sequence was identified as being critical for nucleocapsid binding in an in vitro peptide protection assay. The effects of these mutations on virus assembly and function were determined in both vertebrate and invertebrate cells. Amino acid substitutions (at positions E2: 408, 410, 411, and 413) reduced infectious virus production in a position-dependent fashion but were not efficient in disrupting assembly in mammalian cells. Deletions in the E2 endodomain (delta406-407, delta409-411, and delta414-417) resulted in the failure to assemble virions in mammalian cells. Electron microscopy of BHK cells transfected with these mutants revealed assembly of nucleocapsids that failed to attach to membranes. However, introduction of these deletion mutants into insect cells resulted in the assembly of virus-like particles but no assayable infectivity. These data help define protein interactions critical for virus assembly and suggest a fundamental difference between Sindbis virus assembly in mammalian and insect cells.
Figures


Similar articles
-
Single amino acid insertions at the junction of the sindbis virus E2 transmembrane domain and endodomain disrupt virus envelopment and alter infectivity.J Virol. 2005 Jun;79(12):7682-97. doi: 10.1128/JVI.79.12.7682-7697.2005. J Virol. 2005. PMID: 15919921 Free PMC article.
-
A single deletion in the membrane-proximal region of the Sindbis virus glycoprotein E2 endodomain blocks virus assembly.J Virol. 2000 May;74(9):4220-8. doi: 10.1128/jvi.74.9.4220-4228.2000. J Virol. 2000. PMID: 10756035 Free PMC article.
-
Single-Site Glycoprotein Mutants Inhibit a Late Event in Sindbis Virus Assembly.J Virol. 2016 Aug 26;90(18):8372-80. doi: 10.1128/JVI.00948-16. Print 2016 Sep 15. J Virol. 2016. PMID: 27412592 Free PMC article.
-
Research on basis of reverse genetics system of a Sindbis-like virus XJ-160.Virol J. 2011 Nov 14;8:519. doi: 10.1186/1743-422X-8-519. Virol J. 2011. PMID: 22082202 Free PMC article. Review.
-
The coronavirus E protein: assembly and beyond.Viruses. 2012 Mar;4(3):363-82. doi: 10.3390/v4030363. Epub 2012 Mar 8. Viruses. 2012. PMID: 22590676 Free PMC article. Review.
Cited by
-
A Systematic Review of the Natural Virome of Anopheles Mosquitoes.Viruses. 2018 Apr 25;10(5):222. doi: 10.3390/v10050222. Viruses. 2018. PMID: 29695682 Free PMC article.
-
The structure of Sindbis virus produced from vertebrate and invertebrate hosts as determined by small-angle neutron scattering.J Virol. 2010 May;84(10):5270-6. doi: 10.1128/JVI.00044-10. Epub 2010 Mar 10. J Virol. 2010. PMID: 20219936 Free PMC article.
-
Structural mutants of dengue virus 2 transmembrane domains exhibit host-range phenotype.Virol J. 2011 Jun 9;8:289. doi: 10.1186/1743-422X-8-289. Virol J. 2011. PMID: 21658241 Free PMC article.
-
A structural and functional perspective of alphavirus replication and assembly.Future Microbiol. 2009 Sep;4(7):837-56. doi: 10.2217/fmb.09.59. Future Microbiol. 2009. PMID: 19722838 Free PMC article. Review.
-
Chikungunya virus host range E2 transmembrane deletion mutants induce protective immunity against challenge in C57BL/6J mice.J Virol. 2013 Jun;87(12):6748-57. doi: 10.1128/JVI.03357-12. Epub 2013 Apr 3. J Virol. 2013. PMID: 23552427 Free PMC article.
References
-
- Bretscher, M. S. 1993. Cholesterol and the Golgi apparatus. Science 261:1280-1281. - PubMed
-
- Brown, D. T., and L. D. Condreay. 1986. Replication of alphaviruses in mosquito cells, p. 171-207. In M. J. Schlesinger (ed.), The Togaveriridae and Flaviviridae. Plenum Publishing Corp., New York, N.Y.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

