Compositionally and functionally distinct editosomes in Trypanosoma brucei
- PMID: 16611942
- PMCID: PMC1464856
- DOI: 10.1261/rna.45506
Compositionally and functionally distinct editosomes in Trypanosoma brucei
Abstract
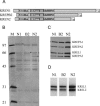
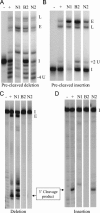
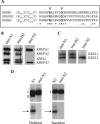
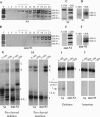
Uridylate insertion/deletion RNA editing in Trypanosoma brucei mitochondria is catalyzed by a multiprotein complex, the approximately 20S editosome. Editosomes purified via three related tagged RNase III proteins, KREN1 (KREPB1/TbMP90), KREPB2 (TbMP67), and KREN2 (KREPB3/TbMP61), had very similar but nonidentical protein compositions, and only the tagged member of these three RNase III proteins was identified in each respective complex. Three new editosome proteins were also identified in these complexes. Each tagged complex catalyzed both precleaved insertion and deletion editing in vitro. However, KREN1 complexes cleaved deletion but not insertion editing sites in vitro, and, conversely, KREN2 complexes cleaved insertion but not deletion editing sites. These specific nuclease activities were abolished by mutations in the putative RNase III catalytic domain of the respective proteins. Thus editosomes appear to be heterogeneous in composition with KREN1 complexes catalyzing cleavage of deletion sites and KREN2 complexes cleaving insertion sites while both can catalyze the U addition, U removal, and ligation steps of editing.
Figures

Similar articles
-
Endonuclease associations with three distinct editosomes in Trypanosoma brucei.J Biol Chem. 2011 Jun 3;286(22):19320-30. doi: 10.1074/jbc.M111.228965. Epub 2011 Apr 7. J Biol Chem. 2011. PMID: 21474442 Free PMC article.
-
A deletion site editing endonuclease in Trypanosoma brucei.Mol Cell. 2005 Nov 11;20(3):403-12. doi: 10.1016/j.molcel.2005.09.016. Mol Cell. 2005. PMID: 16285922
-
An essential RNase III insertion editing endonuclease in Trypanosoma brucei.Proc Natl Acad Sci U S A. 2005 Nov 15;102(46):16614-9. doi: 10.1073/pnas.0506133102. Epub 2005 Nov 3. Proc Natl Acad Sci U S A. 2005. PMID: 16269544 Free PMC article.
-
RNA editing: complexity and complications.Mol Microbiol. 2002 Aug;45(3):591-6. doi: 10.1046/j.1365-2958.2002.03028.x. Mol Microbiol. 2002. PMID: 12139607 Review.
-
'Gestalt,' composition and function of the Trypanosoma brucei editosome.Annu Rev Microbiol. 2012;66:65-82. doi: 10.1146/annurev-micro-092611-150150. Annu Rev Microbiol. 2012. PMID: 22994488 Review.
Cited by
-
Prioritization of Trypanosoma brucei editosome protein interactions interfaces at residue resolution through proteome-scale network analysis.BMC Mol Cell Biol. 2024 Jan 26;25(1):3. doi: 10.1186/s12860-024-00499-4. BMC Mol Cell Biol. 2024. PMID: 38279116 Free PMC article.
-
Editosome accessory factors KREPB9 and KREPB10 in Trypanosoma brucei.Eukaryot Cell. 2012 Jul;11(7):832-43. doi: 10.1128/EC.00046-12. Epub 2012 May 4. Eukaryot Cell. 2012. PMID: 22562468 Free PMC article.
-
An electrochemiluminescent aptamer switch for a high-throughput assay of an RNA editing reaction.RNA. 2009 Oct;15(10):1929-38. doi: 10.1261/rna.1720209. Epub 2009 Aug 20. RNA. 2009. PMID: 19696159 Free PMC article.
-
Trypanosoma brucei RNA editing: coupled cycles of U deletion reveal processive activity of the editing complex.Mol Cell Biol. 2008 Apr;28(7):2437-45. doi: 10.1128/MCB.01886-07. Epub 2008 Jan 28. Mol Cell Biol. 2008. PMID: 18227152 Free PMC article.
-
Guide RNA biogenesis involves a novel RNase III family endoribonuclease in Trypanosoma brucei.RNA. 2011 Oct;17(10):1821-30. doi: 10.1261/rna.2815911. Epub 2011 Aug 2. RNA. 2011. PMID: 21810935 Free PMC article.
References
-
- Aphasizhev R., Sbicego S., Peris M., Jang S.H., Aphasizheva I., Simpson A.M., Rivlin A., Simpson L. Trypanosome mitochondrial 3′ terminal uridylyl transferase (TUTase): The key enzyme in U-insertion/deletion RNA editing. Cell. 2002;108:637–648. - PubMed
-
- Blaszczyk J., Tropea J.E., Bubunenko M., Routzahn K.M., Waugh D.S., Court D.L., Ji X. Crystallographic and modeling studies of RNase III suggest a mechanism for double-stranded RNA cleavage. Structure. 2001;9:1225–1236. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
