The domain of the Bacillus subtilis DEAD-box helicase YxiN that is responsible for specific binding of 23S rRNA has an RNA recognition motif fold
- PMID: 16611943
- PMCID: PMC1464845
- DOI: 10.1261/rna.5906
The domain of the Bacillus subtilis DEAD-box helicase YxiN that is responsible for specific binding of 23S rRNA has an RNA recognition motif fold
Abstract
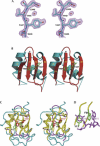
The YxiN protein of Bacillus subtilis is a member of the DbpA subfamily of prokaryotic DEAD-box RNA helicases. Like DbpA, it binds with high affinity and specificity to segments of 23S ribosomal RNA as short as 32 nucleotides (nt) that include hairpin 92. Several experiments have shown that the 76-residue carboxy-terminal domain of YxiN is responsible for the high-affinity RNA binding. The domain has been crystallized and its structure has been solved to 1.7 Angstroms resolution. The structure reveals an RNA recognition motif (RRM) fold that is found in many eukaryotic RNA binding proteins; the RRM fold was not apparent from the amino acid sequence. The domain has two solvent exposed aromatic residues at sites that correspond to the aromatic residues of the ribonucleoprotein (RNP) motifs RNP1 and RNP2 that are essential for RNA binding in many RRMs. However, mutagenesis of these residues (Tyr404 and Tyr447) to alanine has little effect on RNA affinity, suggesting that the YxiN domain binds target RNAs in a manner that differs from the binding mode commonly found in many eukaryotic RRMs.
Figures

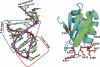
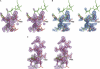
Similar articles
-
The carboxy-terminal domain of the DExDH protein YxiN is sufficient to confer specificity for 23S rRNA.J Mol Biol. 2002 Dec 6;324(4):625-36. doi: 10.1016/s0022-2836(02)01140-3. J Mol Biol. 2002. PMID: 12460566
-
Structure of the RNA binding domain of a DEAD-box helicase bound to its ribosomal RNA target reveals a novel mode of recognition by an RNA recognition motif.J Mol Biol. 2010 Sep 17;402(2):412-27. doi: 10.1016/j.jmb.2010.07.040. Epub 2010 Jul 29. J Mol Biol. 2010. PMID: 20673833 Free PMC article.
-
Cloning and biochemical characterization of Bacillus subtilis YxiN, a DEAD protein specifically activated by 23S rRNA: delineation of a novel sub-family of bacterial DEAD proteins.Nucleic Acids Res. 1999 Oct 1;27(19):3811-20. doi: 10.1093/nar/27.19.3811. Nucleic Acids Res. 1999. PMID: 10481020 Free PMC article.
-
RNA-protein complexes.Curr Opin Struct Biol. 1996 Feb;6(1):53-61. doi: 10.1016/s0959-440x(96)80095-9. Curr Opin Struct Biol. 1996. PMID: 8696973 Review.
-
RNA sculpting by the primordial Helix-clasp-Helix-Strand-Loop (HcH-SL) motif enforces chemical recognition enabling diverse KH domain functions.J Biol Chem. 2025 May;301(5):108474. doi: 10.1016/j.jbc.2025.108474. Epub 2025 Apr 2. J Biol Chem. 2025. PMID: 40185232 Free PMC article. Review.
Cited by
-
DEAD-box helicases as integrators of RNA, nucleotide and protein binding.Biochim Biophys Acta. 2013 Aug;1829(8):884-93. doi: 10.1016/j.bbagrm.2013.02.002. Epub 2013 Feb 15. Biochim Biophys Acta. 2013. PMID: 23416748 Free PMC article. Review.
-
Dead-box proteins: a family affair--active and passive players in RNP-remodeling.Nucleic Acids Res. 2006;34(15):4168-80. doi: 10.1093/nar/gkl468. Epub 2006 Aug 26. Nucleic Acids Res. 2006. PMID: 16936318 Free PMC article. Review.
-
The putative RNase P motif in the DEAD box helicase Hera is dispensable for efficient interaction with RNA and helicase activity.Nucleic Acids Res. 2008 Oct;36(18):5800-11. doi: 10.1093/nar/gkn581. Epub 2008 Sep 9. Nucleic Acids Res. 2008. PMID: 18782831 Free PMC article.
-
Mutation of the arginine finger in the active site of Escherichia coli DbpA abolishes ATPase and helicase activity and confers a dominant slow growth phenotype.Nucleic Acids Res. 2008 Jan;36(1):41-50. doi: 10.1093/nar/gkm926. Epub 2007 Nov 5. Nucleic Acids Res. 2008. PMID: 17986459 Free PMC article.
-
DbpA is a region-specific RNA helicase.Biopolymers. 2017 Mar;107(3):10.1002/bip.23001. doi: 10.1002/bip.23001. Biopolymers. 2017. PMID: 27813083 Free PMC article.
References
-
- Allain F.H., Gilbert D.E., Bouvet P., Feigon J. Solution structure of the two N-terminal RNA-binding domains of nucleolin and NMR study of the interaction with its RNA target. J. Mol. Biol. 2000b;303:227–241. - PubMed
-
- Brünger A.T., Adams P.D., Clore G.M., DeLano W.L., Gros P., Grosse-Kunstleve R.W., Jiang J.S., Kuszewski J., Nilges M., Pannu N.S., et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. - PubMed
-
- Caruthers J.M., McKay D.B. Helicase structure and mechanism. Curr. Opin. Struct. Biol. 2002;12:123–133. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous
