Role for Upf2p phosphorylation in Saccharomyces cerevisiae nonsense-mediated mRNA decay
- PMID: 16611983
- PMCID: PMC1447418
- DOI: 10.1128/MCB.26.9.3390-3400.2006
Role for Upf2p phosphorylation in Saccharomyces cerevisiae nonsense-mediated mRNA decay
Abstract
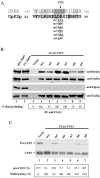
Premature termination (nonsense) codons trigger rapid mRNA decay by the nonsense-mediated mRNA decay (NMD) pathway. Two conserved proteins essential for NMD, UPF1 and UPF2, are phosphorylated in higher eukaryotes. The phosphorylation and dephosphorylation of UPF1 appear to be crucial for NMD, as blockade of either event in Caenorhabditis elegans and mammals largely prevents NMD. The universality of this phosphorylation/dephosphorylation cycle pathway has been questioned, however, because the well-studied Saccharomyces cerevisiae NMD pathway has not been shown to be regulated by phosphorylation. Here, we used in vitro and in vivo biochemical techniques to show that both S. cerevisiae Upf1p and Upf2p are phosphoproteins. We provide evidence that the phosphorylation of the N-terminal region of Upf2p is crucial for its interaction with Hrp1p, an RNA-binding protein that we previously showed is essential for NMD. We identify specific amino acids in Upf2p's N-terminal domain, including phosphorylated serines, which dictate both its interaction with Hrp1p and its ability to elicit NMD. Our results indicate that phosphorylation of UPF1 and UPF2 is a conserved event in eukaryotes and for the first time provide evidence that Upf2p phosphorylation is crucial for NMD.
Figures




Similar articles
-
Inactivation of NMD increases viability of sup45 nonsense mutants in Saccharomyces cerevisiae.BMC Mol Biol. 2007 Aug 16;8:71. doi: 10.1186/1471-2199-8-71. BMC Mol Biol. 2007. PMID: 17705828 Free PMC article.
-
Genetic background affects relative nonsense mRNA accumulation in wild-type and upf mutant yeast strains.Curr Genet. 2003 Jun;43(3):171-7. doi: 10.1007/s00294-003-0386-3. Epub 2003 Apr 15. Curr Genet. 2003. PMID: 12695845
-
Novel Upf2p orthologues suggest a functional link between translation initiation and nonsense surveillance complexes.Mol Cell Biol. 2000 Dec;20(23):8944-57. doi: 10.1128/MCB.20.23.8944-8957.2000. Mol Cell Biol. 2000. PMID: 11073994 Free PMC article.
-
NMD monitors translational fidelity 24/7.Curr Genet. 2017 Dec;63(6):1007-1010. doi: 10.1007/s00294-017-0709-4. Epub 2017 May 23. Curr Genet. 2017. PMID: 28536849 Free PMC article. Review.
-
Defining nonsense-mediated mRNA decay intermediates in human cells.Methods. 2019 Feb 15;155:68-76. doi: 10.1016/j.ymeth.2018.12.005. Epub 2018 Dec 19. Methods. 2019. PMID: 30576707 Free PMC article. Review.
Cited by
-
Dbp5/DDX19 between Translational Readthrough and Nonsense Mediated Decay.Int J Mol Sci. 2020 Feb 6;21(3):1085. doi: 10.3390/ijms21031085. Int J Mol Sci. 2020. PMID: 32041247 Free PMC article. Review.
-
Upf1 potentially serves as a RING-related E3 ubiquitin ligase via its association with Upf3 in yeast.RNA. 2008 Sep;14(9):1950-8. doi: 10.1261/rna.536308. Epub 2008 Aug 1. RNA. 2008. PMID: 18676617 Free PMC article.
-
Mechanism, factors, and physiological role of nonsense-mediated mRNA decay.Cell Mol Life Sci. 2015 Dec;72(23):4523-44. doi: 10.1007/s00018-015-2017-9. Epub 2015 Aug 18. Cell Mol Life Sci. 2015. PMID: 26283621 Free PMC article. Review.
-
The shuttling protein Npl3 promotes translation termination accuracy in Saccharomyces cerevisiae.J Mol Biol. 2009 Dec 4;394(3):410-22. doi: 10.1016/j.jmb.2009.08.067. Epub 2009 Sep 3. J Mol Biol. 2009. PMID: 19733178 Free PMC article.
-
A highly conserved region essential for NMD in the Upf2 N-terminal domain.J Mol Biol. 2014 Nov 11;426(22):3689-3702. doi: 10.1016/j.jmb.2014.09.015. Epub 2014 Sep 30. J Mol Biol. 2014. PMID: 25277656 Free PMC article.
References
-
- Amrani, N., R. Ganesan, S. Kervestin, D. A. Mangus, S. Ghosh, and A. Jacobson. 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432:112-118. - PubMed
-
- Baker, K. E., and R. Parker. 2004. Nonsense-mediated mRNA decay: terminating erroneous gene expression. Curr. Opin. Cell Biol. 16:293-299. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
