Development and implementation of an algorithm for detection of protein complexes in large interaction networks
- PMID: 16613608
- PMCID: PMC1473204
- DOI: 10.1186/1471-2105-7-207
Development and implementation of an algorithm for detection of protein complexes in large interaction networks
Abstract
Background: After complete sequencing of a number of genomes the focus has now turned to proteomics. Advanced proteomics technologies such as two-hybrid assay, mass spectrometry etc. are producing huge data sets of protein-protein interactions which can be portrayed as networks, and one of the burning issues is to find protein complexes in such networks. The enormous size of protein-protein interaction (PPI) networks warrants development of efficient computational methods for extraction of significant complexes.
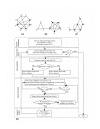
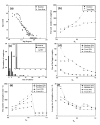
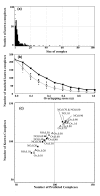
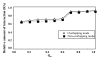
Results: This paper presents an algorithm for detection of protein complexes in large interaction networks. In a PPI network, a node represents a protein and an edge represents an interaction. The input to the algorithm is the associated matrix of an interaction network and the outputs are protein complexes. The complexes are determined by way of finding clusters, i. e. the densely connected regions in the network. We also show and analyze some protein complexes generated by the proposed algorithm from typical PPI networks of Escherichia coli and Saccharomyces cerevisiae. A comparison between a PPI and a random network is also performed in the context of the proposed algorithm.
Conclusion: The proposed algorithm makes it possible to detect clusters of proteins in PPI networks which mostly represent molecular biological functional units. Therefore, protein complexes determined solely based on interaction data can help us to predict the functions of proteins, and they are also useful to understand and explain certain biological processes.
Figures



References
-
- Gavin AC, Bösche M, Krause R, Grandi P, Marzioch M, Bauer A, Schultz J, Rick JM, Michon AM, Cruciat CM, Remor M, Höfert C, Schelder M, Brajenovic M, Ruffner H, Merino A, Klein K, Hudak M, Dickson D, Rudi T, Gnau V, Bauch A, Bastuck S, Huhse B, Leutwein C, Heurtier MA, Copley RR, Edelmann A, Querfurth E, Rybin V, Drewes G, Raida M, Bouwmeester T, Bork P, Seraphin B, Kuster B, Neubauer G, Superti-Furga G. Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature. 2002;415:141–147. doi: 10.1038/415141a. - DOI - PubMed
-
- Ho Y, Gruhler A, Heilbut A, Bader GD, Moore L, Adams SL, Millar A, Taylor P, Bennett K, Boutilier K, Yang L, Wolting C, Donaldson I, Schandorff S, Shewnarane J, Vo M, Taggart J, Goudreault M, Muskat B, Alfarano C, Dewar D, Lin Z, Michalickova K, Willems AR, Sassi H, Nielsen PA, Rasmussen KJ, Andersen JR, Johansen LE, Hansen LH, Jespersen H, Podtelejnikov A, Nielsen E, Crawford J, Poulsen V, Sorensen BD, Matthiesen J, Hendrickson RC, Gleeson F, Pawson T, Moran MF, Durocher D, Mann M, Hogue CW, Figeys D, Tyers M. Systematic identification of protein complexes in Saccharomyces cerevisiae by mass spectrometry. Nature. 2002;415:180–183. doi: 10.1038/415180a. - DOI - PubMed
-
- Seidman SB. Network structure and minimum degree. Social Networks. 1983;5:269–287. doi: 10.1016/0378-8733(83)90028-X. - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

