Corto and DSP1 interact and bind to a maintenance element of the Scr Hox gene: understanding the role of Enhancers of trithorax and Polycomb
- PMID: 16613610
- PMCID: PMC1459216
- DOI: 10.1186/1741-7007-4-9
Corto and DSP1 interact and bind to a maintenance element of the Scr Hox gene: understanding the role of Enhancers of trithorax and Polycomb
Abstract
Background: Polycomb-group genes (PcG) encode proteins that maintain homeotic (Hox) gene repression throughout development. Conversely, trithorax-group (trxG) genes encode positive factors required for maintenance of long term Hox gene activation. Both kinds of factors bind chromatin regions called maintenance elements (ME). Our previous work has shown that corto, which codes for a chromodomain protein, and dsp1, which codes for an HMGB protein, belong to a class of genes called the Enhancers of trithorax and Polycomb (ETP) that interact with both PcG and trxG. Moreover, dsp1 interacts with the Hox gene Scr, the DSP1 protein is present on a Scr ME in S2 cells but not in embryos. To understand better the role of ETP, we addressed genetic and molecular interactions between corto and dsp1.
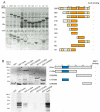
Results: We show that Corto and DSP1 proteins co-localize at 91 sites on polytene chromosomes and co-immunoprecipitate in embryos. They interact directly through the DSP1 HMG-boxes and the amino-part of Corto, which contains a chromodomain. In order to search for a common target, we performed a genetic interaction analysis. We observed that corto mutants suppressed dsp11 sex comb phenotypes and enhanced AntpScx phenotypes, suggesting that corto and dsp1 are simultaneously involved in the regulation of Scr. Using chromatin immunoprecipitation of the Scr ME, we found that Corto was present on this ME both in Drosophila S2 cells and in embryos, whereas DSP1 was present only in S2 cells.
Conclusion: Our results reveal that the proteins Corto and DSP1 are differently recruited to a Scr ME depending on whether the ME is active, as seen in S2 cells, or inactive, as in most embryonic cells. The presence of a given combination of ETPs on an ME would control the recruitment of either PcG or TrxG complexes, propagating the silenced or active state.
Figures


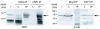

References
-
- Ingham PW. A clonal analysis of the requirement for the trithorax gene in the diversification of segments in Drosophila. J Embryol Exp Morphol. 1985;89:349–365. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

