Comparison of characteristics and function of translation termination signals between and within prokaryotic and eukaryotic organisms
- PMID: 16614446
- PMCID: PMC1435984
- DOI: 10.1093/nar/gkl074
Comparison of characteristics and function of translation termination signals between and within prokaryotic and eukaryotic organisms
Abstract
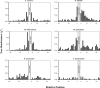
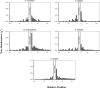
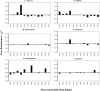
Six diverse prokaryotic and five eukaryotic genomes were compared to deduce whether the protein synthesis termination signal has common determinants within and across both kingdoms. Four of the six prokaryotic and all of the eukaryotic genomes investigated demonstrated a similar pattern of nucleotide bias both 5' and 3' of the stop codon. A preferred core signal of 4 nt was evident, encompassing the stop codon and the following nucleotide. Codons decoded by hyper-modified tRNAs were over-represented in the region 5' to the stop codon in genes from both kingdoms. The origin of the 3' bias was more variable particularly among the prokaryotic organisms. In both kingdoms, genes with the highest expression index exhibited a strong bias but genes with the lowest expression showed none. Absence of bias in parasitic prokaryotes may reflect an absence of pressure to evolve more efficient translation. Experiments were undertaken to determine if a correlation existed between bias in signal abundance and termination efficiency. In Escherichia coli signal abundance correlated with termination efficiency for UAA and UGA stop codons, but not in mammalian cells. Termination signals that were highly inefficient could be made more efficient by increasing the concentration of the cognate decoding release factor.
Figures


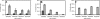

References
-
- Poole E., Tate W. Release factors and their role as decoding proteins: specificity and fidelity for termination of protein synthesis. Biochim. Biophys. Acta. 2000;1493:1–11. - PubMed
-
- Nakamura Y., Ito K. Making sense of mimic in translation termination. Trends Biochem. Sci. 2003;28:99–105. - PubMed
-
- Buckingham R.H., Grentzmann G., Kisselev L. Polypeptide chain release factors. Mol. Microbiol. 1997;24:449–456. - PubMed
-
- Inge-Vechtomov S., Zhouravleva G., Philippe M. Eukaryotic release factors (eRFs) history. Biol. Cell. 2003;95:195–109. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

