Magnesium-induced assembly of a complete DNA polymerase catalytic complex
- PMID: 16615916
- PMCID: PMC1868394
- DOI: 10.1016/j.str.2006.01.011
Magnesium-induced assembly of a complete DNA polymerase catalytic complex
Abstract
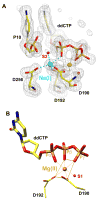
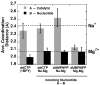
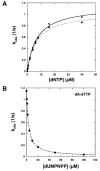
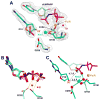
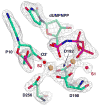
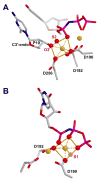
The molecular details of the nucleotidyl transferase reaction have remained speculative, as strategies to trap catalytic intermediates for structure determination utilize substrates lacking the primer terminus 3'-OH and catalytic Mg2+, resulting in an incomplete and distorted active site geometry. Since the geometric arrangement of these essential atoms will impact chemistry, structural insight into fidelity strategies has been hampered. Here, we present a crystal structure of a precatalytic complex of a DNA polymerase with bound substrates that include the primer 3'-OH and catalytic Mg2+. This catalytic intermediate was trapped with a nonhydrolyzable deoxynucleotide analog. Comparison with two new structures of DNA polymerase beta lacking the 3'-OH or catalytic Mg2+ is described. These structures provide direct evidence that both atoms are required to achieve a proper geometry necessary for an in-line nucleophilic attack of O3' on the alphaP of the incoming nucleotide.
Figures
References
-
- Arndt JW, Gong W, Zhong X, Showalter AK, Liu J, Dunlap CA, Lin Z, Paxson C, Tsai MD, Chan MK. Insight into the catalytic mechanism of DNA polymerase β: Structures of intermediate complexes. Biochemistry. 2001;40:5368–5375. - PubMed
-
- Batra VK, Beard WA, Shock DD, Pedersen LC, Wilson SH. Nucleotide-induced DNA polymerase active site motions accomodating a mutagenic DNA intermediate. Structure (Camb) 2005;13:1225–1233. - PubMed
-
- Beard WA, Wilson SH. Purification and domain-mapping of mammalian DNA polymerase β. Methods Enzymol. 1995;262:98–107. - PubMed
-
- Beard WA, Wilson SH. Structural insights into DNA polymerase β fidelity: Hold tight if you want it right. Chem Biol. 1998;5:R7–R13. - PubMed
-
- Beard WA, Wilson SH. Structural design of a eukaryotic DNA repair polymerase: DNA polymerase β. Mutat Res. 2000;460:231–244. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

