MTA1, a transcriptional activator of breast cancer amplified sequence 3
- PMID: 16617102
- PMCID: PMC1458939
- DOI: 10.1073/pnas.0601989103
MTA1, a transcriptional activator of breast cancer amplified sequence 3
Erratum in
- Proc Natl Acad Sci U S A. 2013 Mar 5;110(10):4147-8
Abstract
Here we define a function of metastasis-associated protein 1 (MTA1), a presumed corepressor of estrogen receptor alpha (ERalpha), as a transcriptional activator of Breast Cancer Amplified Sequence 3 (BCAS3), a gene amplified and overexpressed in breast cancers. We identified BCAS3 as a MTA1 chromatin target in a functional genomic screen. MTA1 stimulation of BCAS3 transcription required ERalpha and involved a functional ERE half-site in BCAS3. Furthermore, we discovered that MTA1 is acetylated on lysine 626, and that this acetylation is necessary for a productive transcriptional recruitment of RNA polymerase II complex to the BCAS3 enhancer sequence. BCAS3 expression was elevated in mammary tumors from MTA1 transgenic mice and 60% of the human breast tumors, and correlated with the coexpression of MTA1 as well as with tumor grade and proliferation of primary breast tumor samples. These findings reveal a previously unrecognized function of MTA1 in stimulating BCAS3 expression and suggest an important role for MTA1-BCAS3 pathway in promoting cancerous phenotypes in breast tumor cells.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


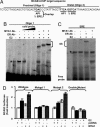

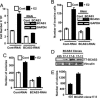

References
-
- Fowler A. M., Alarid E. T. Sci. STKE. 2004;2004:e51.
-
- McKenna N. J., O'Malley B. W. Cell. 2002;108:465–474. - PubMed
-
- McKenna N. J., Lanz R. B., O'Malley B. W. Endocr. Rev. 1999;20:321–344. - PubMed
-
- Toh Y., Pencil S. D., Nicolson G. L. J. Biol. Chem. 1994;269:22958–22963. - PubMed
-
- Bowen N. J., Fujita N., Kajita M., Wade P. A. Biochim. Biophys. Acta. 2004;1677:52–57. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

