Dual roles for the trimeric G protein Go in asymmetric cell division in Drosophila
- PMID: 16617104
- PMCID: PMC1436022
- DOI: 10.1073/pnas.0601853103
Dual roles for the trimeric G protein Go in asymmetric cell division in Drosophila
Abstract
During asymmetric division, a cell polarizes and differentially distributes components to its opposite ends. The subsequent division differentially segregates the two component pools to the daughters, which thereby inherit different developmental directives. In Drosophila sensory organ precursor cells, the localization of Numb protein to the cell's anterior cortex is a key patterning event and is achieved by the combined action of many proteins, including Pins, which itself is localized anteriorly. Here, a role is described for the trimeric G protein Go in the anterior localization of Numb and daughter cell fate specification. Go is shown to interact with Pins. In addition to a role in recruiting Numb to an asymmetric location in the cell's cortex, Go transduces a signal from the Frizzled receptor that directs the position in which the complex forms. Thus, Go likely integrates the signaling that directs the formation of the complex with the signaling that directs where the complex forms.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


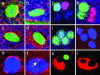
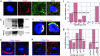


References
-
- Horvitz H. R., Herskowitz I. Cell. 1992;68:237–255. - PubMed
-
- Van Haastert P. J., Devreotes P. N. Nat. Rev. Mol. Cell Biol. 2004;5:626–634. - PubMed
-
- Huber A. B., Kolodkin A. L., Ginty D. D., Cloutier J. F. Annu. Rev. Neurosci. 2003;26:509–563. - PubMed
-
- Chant J. Annu. Rev. Cell Dev. Biol. 1999;15:365–391. - PubMed
-
- Adler P. N. Dev. Cell. 2002;2:525–535. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

