Evidence for a role of developmental genes in the origin of obesity and body fat distribution
- PMID: 16617105
- PMCID: PMC1458940
- DOI: 10.1073/pnas.0601752103
Evidence for a role of developmental genes in the origin of obesity and body fat distribution
Abstract
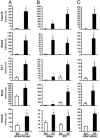
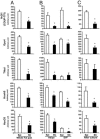
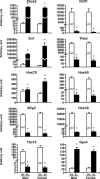
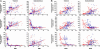
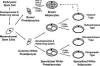
Obesity, especially central obesity, is a hereditable trait associated with a high risk for development of diabetes and metabolic disorders. Combined gene expression analysis of adipocyte- and preadipocyte-containing fractions from intraabdominal and subcutaneous adipose tissue of mice revealed coordinated depot-specific differences in expression of multiple genes involved in embryonic development and pattern specification. These differences were intrinsic and persisted during in vitro culture and differentiation. Similar depot-specific differences in expression of developmental genes were observed in human subcutaneous versus visceral adipose tissue. Furthermore, in humans, several genes exhibited changes in expression that correlated closely with body mass index and/or waist/hip ratio. Together, these data suggest that genetically programmed developmental differences in adipocytes and their precursors in different regions of the body play an important role in obesity, body fat distribution, and potential functional differences between internal and subcutaneous adipose tissue.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
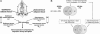
References
-
- Lean M. E. Proc. Nutr. Soc.; 2000. pp. 331–336. - PubMed
-
- Mauriege P., Despres J. P., Moorjani S., Prud’Homme D., Lamarche B., Bouchard C., Nadeau A., Tremblay A., Lupien P. J. Eur. J. Clin. Invest. 1993;23:729–740. - PubMed
-
- Gillum R. F. J. Chronic Dis. 1987;40:421–428. - PubMed
-
- Kissebah A. H., Krakower G. R. Physiol. Rev. 1994;74:761–811. - PubMed
-
- Abate N., Garg A. Prog. Lipid Res. 1995;34:53–70. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

