EM measurements define the dimensions of the "30-nm" chromatin fiber: evidence for a compact, interdigitated structure
- PMID: 16617109
- PMCID: PMC1436021
- DOI: 10.1073/pnas.0601212103
EM measurements define the dimensions of the "30-nm" chromatin fiber: evidence for a compact, interdigitated structure
Abstract
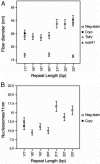
Chromatin structure plays a fundamental role in the regulation of nuclear processes such as DNA transcription, replication, recombination, and repair. Despite considerable efforts during three decades, the structure of the 30-nm chromatin fiber remains controversial. To define fiber dimensions accurately, we have produced very long and regularly folded 30-nm fibers from in vitro reconstituted nucleosome arrays containing the linker histone and with increasing nucleosome repeat lengths (10 to 70 bp of linker DNA). EM measurements show that the dimensions of these fully folded fibers do not increase linearly with increasing linker length, a finding that is inconsistent with two-start helix models. Instead, we find that there are two distinct classes of fiber structure, both with unexpectedly high nucleosome density: arrays with 10 to 40 bp of linker DNA all produce fibers with a diameter of 33 nm and 11 nucleosomes per 11 nm, whereas arrays with 50 to 70 bp of linker DNA all produce 44-nm-wide fibers with 15 nucleosomes per 11 nm. Using the physical constraints imposed by these measurements, we have built a model in which tight nucleosome packing is achieved through the interdigitation of nucleosomes from adjacent helical gyres. Importantly, the model closely matches raw image projections of folded chromatin arrays recorded in the solution state by using electron cryo-microscopy.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures



References
-
- Davies H. G., Small J. V. Nature. 1968;217:1122–1125. - PubMed
-
- Ris H., Kubai D. F. Annu. Rev. Genet. 1970;4:263–294. - PubMed
-
- Finch J. T., Lutter L. C., Rhodes D., Brown R. S., Rushton B., Levitt M., Klug A. Nature. 1977;269:29–36. - PubMed
-
- Richmond T. J., Finch J. T., Rushton B., Rhodes D., Klug A. Nature. 1984;311:532–537. - PubMed
-
- Luger K., Mader A. W., Richmond R. K., Sargent D. F., Richmond T. J. Nature. 1997;389:251–260. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

