Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation
- PMID: 16618808
- PMCID: PMC1472299
- DOI: 10.1101/gad.1406006
Cross-regulation between Notch and p63 in keratinocyte commitment to differentiation
Abstract
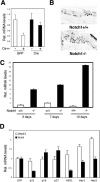
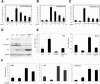
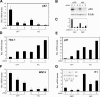
Notch signaling promotes commitment of keratinocytes to differentiation and suppresses tumorigenesis. p63, a p53 family member, has been implicated in establishment of the keratinocyte cell fate and/or maintenance of epithelial self-renewal. Here we show that p63 expression is suppressed by Notch1 activation in both mouse and human keratinocytes through a mechanism independent of cell cycle withdrawal and requiring down-modulation of selected interferon-responsive genes, including IRF7 and/or IRF3. In turn, elevated p63 expression counteracts the ability of Notch1 to restrict growth and promote differentiation. p63 functions as a selective modulator of Notch1-dependent transcription and function, with the Hes-1 gene as one of its direct negative targets. Thus, a complex cross-talk between Notch and p63 is involved in the balance between keratinocyte self-renewal and differentiation.
Figures






References
-
- Artavanis-Tsakonas S., Rand M.D., Lake R.J. Notch signaling: Cell fate control and signal integration in development. Science. 1999;284:770–776. - PubMed
-
- Barbieri C.E., Perez C.A., Johnson K.N., Ely K.A., Billheimer D., Pietenpol J.A. IGFBP-3 is a direct target of transcriptional regulation by ΔNp63α in squamous epithelium. Cancer Res. 2005;65:2314–2320. - PubMed
-
- Bernassola F., Oberst A., Melino G., Pandolfi P.P. The promyelocytic leukaemia protein tumour suppressor functions as a transcriptional regulator of p63. Oncogene. 2005;24:6982–6986. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
