Crystal structures of gamma-glutamyltranspeptidase from Escherichia coli, a key enzyme in glutathione metabolism, and its reaction intermediate
- PMID: 16618936
- PMCID: PMC1458908
- DOI: 10.1073/pnas.0511020103
Crystal structures of gamma-glutamyltranspeptidase from Escherichia coli, a key enzyme in glutathione metabolism, and its reaction intermediate
Abstract
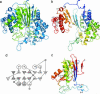
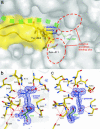
Gamma-glutamyltranspeptidase (GGT) is a heterodimic enzyme that is generated from the precursor protein through posttranslational processing and catalyzes the hydrolysis of gamma-glutamyl bonds in gamma-glutamyl compounds such as glutathione and/or the transfer of the gamma-glutamyl group to other amino acids and peptides. We have determined the crystal structure of GGT from Escherichia coli K-12 at 1.95 A resolution. GGT has a stacked alphabetabetaalpha fold comprising the large and small subunits, similar to the folds seen in members of the N-terminal nucleophile hydrolase superfamily. The active site Thr-391, the N-terminal residue of the small subunit, is located in the groove, from which the pocket for gamma-glutamyl moiety binding follows. We have further determined the structure of the gamma-glutamyl-enzyme intermediate trapped by flash cooling the GGT crystal soaked in glutathione solution and the structure of GGT in complex with l-glutamate. These structures revealed how the gamma-glutamyl moiety and l-glutamate are recognized by the enzyme. A water molecule was seen on the carbonyl carbon of the gamma-glutamyl-Thr-391 Ogamma bond in the intermediate that is to be hydrolyzed. Notably the residues essential for GGT activity (Arg-114, Asp-433, Ser-462, and Ser-463 in E. coli GGT) shown by site-directed mutagenesis of human GGT are all involved in the binding of the gamma-glutamyl moiety. The structure of E. coli GGT presented here, together with sequence alignment of GGTs, may be applicable to interpret the biochemical and genetic data of other GGTs.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

