Archaeology and evolution of transfer RNA genes in the Escherichia coli genome
- PMID: 16618964
- PMCID: PMC1464854
- DOI: 10.1261/rna.2272306
Archaeology and evolution of transfer RNA genes in the Escherichia coli genome
Abstract
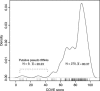
Transfer RNA genes tend to be presented in multiple copies in the genomes of most organisms, from bacteria to eukaryotes. The evolution and genomic structure of tRNA genes has been a somewhat neglected area of molecular evolution. Escherichia coli, the first phylogenetic species for which more than two different strains have been sequenced, provides an invaluable framework to study the evolution of tRNA genes. In this work, a detailed analysis of the tRNA structure of the genomes of Escherichia coli strains K12, CFT073, and O157:H7, Shigella flexneri 2a 301, and Salmonella typhimurium LT2 was carried out. A phylogenetic analysis of these organisms was completed, and an archaeological map depicting the main events in the evolution of tRNA genes was drawn. It is shown that duplications, deletions, and horizontal gene transfers are the main factors driving tRNA evolution in these genomes. On average, 0.64 tRNA insertions/duplications occur every million years (Myr) per genome per lineage, while deletions occur at the slower rate of 0.30 per million years per genome per lineage. This work provides a first genomic glance at the problem of tRNA evolution as a repetitive process, and the relationship of this mechanism to genome evolution and codon usage is discussed.
Figures


References
-
- Bachellier S., Gilson E., Hofnung M., Hill C.W. Repeated sequences. In: Neidhardt F.C., editor. Escherichia coli and Salmonella: Cellular and molecular biology. ASM Press; Washington, DC: 1996. pp. 2708–2720.
-
- Bennetzen J.L., Hall B.D. Codon selection in yeast. J. Biol. Chem. 1982;257:3026–3031. - PubMed
-
- Blattner F.R., Plunkett G., 3rd, Bloch C.A., Perna N.T., Burland V., Riley M., Collado-Vides J., Glasner J.D., Rode C.K., Mayhew G.F., et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1474. - PubMed
-
- Bulmer M. Coevolution of codon usage and transfer RNA abundance. Nature. 1987;325:728–730. - PubMed
-
- Charlesworth B., Sniegowski P., Stephan W. The evolutionary dynamics of repetitive DNA in eukaryotes. Nature. 1994;371:215–220. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
