Functional characterization of the conserved amino acids in Pop1p, the largest common protein subunit of yeast RNases P and MRP
- PMID: 16618965
- PMCID: PMC1464857
- DOI: 10.1261/rna.23206
Functional characterization of the conserved amino acids in Pop1p, the largest common protein subunit of yeast RNases P and MRP
Abstract
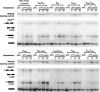
RNase P and RNase MRP are ribonucleoprotein enzymes required for 5'-end maturation of precursor tRNAs (pre-tRNAs) and processing of precursor ribosomal RNAs, respectively. In yeast, RNase P and MRP holoenzymes have eight protein subunits in common, with Pop1p being the largest at >100 kDa. Little is known about the functions of Pop1p, beyond the fact that it binds specifically to the RNase P RNA subunit, RPR1 RNA. In this study, we refined the previous Pop1 phylogenetic sequence alignment and found four conserved regions. Highly conserved amino acids in yeast Pop1p were mutagenized by randomization and conditionally defective mutations were obtained. Effects of the Pop1p mutations on pre-tRNA processing, pre-rRNA processing, and stability of the RNA subunits of RNase P and MRP were examined. In most cases, functional defects in RNase P and RNase MRP in vivo were consistent with assembly defects of the holoenzymes, although moderate kinetic defects in RNase P were also observed. Most mutations affected both pre-tRNA and pre-rRNA processing, but a few mutations preferentially interfered with only RNase P or only RNase MRP. In addition, one temperature-sensitive mutation had no effect on either tRNA or rRNA processing, consistent with an additional role for RNase P, RNase MRP, or Pop1p in some other form. This study shows that the Pop1p subunit plays multiple roles in the assembly and function of of RNases P and MRP, and that the functions can be differentiated through the mutations in conserved residues.
Figures







References
-
- Aravind L., Koonin E.V. G-patch: A new conserved domain in eukaryotic RNA-processing proteins and type D retroviral polyproteins. Trends Biochem. Sci. 1999;24:342–344. - PubMed
-
- Beebe J.A., Fierke C.A. A kinetic mechanism for cleavage of precursor tRNA(Asp) catalyzed by the RNA component of Bacillus subtilis ribonuclease P. Biochemistry. 1994;33:10294–10304. - PubMed
-
- Brusca E.M., True H.L., Celander D.W. Novel RNA-binding properties of Pop3p support a role for eukaryotic RNase P protein subunits in substrate recognition. J. Biol. Chem. 2001;276:42543–42548. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
