Residues in two homology blocks on the amino side of the tRNase Z His domain contribute unexpectedly to pre-tRNA 3' end processing
- PMID: 16618969
- PMCID: PMC1464858
- DOI: 10.1261/rna.4206
Residues in two homology blocks on the amino side of the tRNase Z His domain contribute unexpectedly to pre-tRNA 3' end processing
Abstract
tRNase Z, which can endonucleolytically remove pre-tRNA 3'-end trailers, possesses the signature His domain (HxHxDH; Motif II) of the beta-lactamase family of metal-dependent hydrolases. Motif II combines with Motifs III-V on its carboxy side to coordinate two divalent metal ions, constituting the catalytic core. The PxKxRN loop and Motif I on the amino side of Motif II have been suggested to modulate tRNase Z activity, including the anti-determinant effect of CCA in mature tRNA. Ala walks through these two homology blocks reveal residues in which the substitutions unexpectedly reduce catalytic efficiency. While substitutions in Motif II can drastically affect k(cat) without affecting k(M), five- to 15-fold increases in k(M) are observed with substitutions in several conserved residues in the PxKxRN loop and Motif I. These increases in k(M) suggest a model for substrate binding. Expressed tRNase Z processes mature tRNA with CCA at the 3' end approximately 80 times less efficiently than a pre-tRNA possessing natural sequence of the 3'-end trailer, due to reduced k(cat) with no effect on k(M), showing the CCA anti-determinant to be a characteristic of this enzyme.
Figures




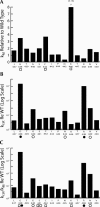
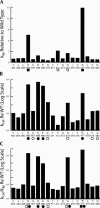

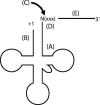
References
-
- Aebi M., Kirchner G., Chen J.Y., Vijayraghavan U., Jacobson A., Martin N.C., Abelson J. Isolation of a temperature-sensitive mutant with an altered tRNA nucleotidyltransferase and cloning of the gene encoding tRNA nucleotidyltransferase in the yeast Saccharomyces cerevisiae . J. Biol. Chem. 1990;265:16216–16220. - PubMed
-
- Baillat D., Hakimi M.A., Naar A.M., Shilatifard A., Cooch N., Shiekhattar R. Integrator, a multiprotein mediator of small nuclear RNA processing, associates with the C-terminal repeat of RNA polymerase II. Cell. 2005;123:265–276. - PubMed
-
- Castaño J.G., Tobian J.A., Zasloff M. Purification and characterization of an endonuclease from Xenopus laevis ovaries which accurately processes the 3′ terminus of human pre-tRNA-Met(i) (3′ pre-tRNase) J. Biol. Chem. 1985;260:9002–9008. - PubMed
-
- Chen J.Y., Martin N.C. Biosynthesis of tRNA in yeast mitochondria. An endonuclease is responsible for the 3′-processing of tRNA precursors. J. Biol. Chem. 1988;263:13677–13682. - PubMed
-
- de la Sierra-Gallay I.L., Pellegrini O., Condon C. Structural basis for substrate binding, cleavage and allostery in the tRNA maturase RNase Z. Nature. 2005;433:657–661. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous
