Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter
- PMID: 16620112
- PMCID: PMC2580050
- DOI: 10.1021/ja0563861
Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter
Abstract
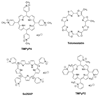
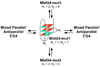
The human bcl-2 gene contains a GC-rich region upstream of the P1 promoter that has been shown to be critically involved in the regulation of bcl-2 gene expression. We have demonstrated that the guanine-rich strand of the DNA in this region can form any one of three distinct intramolecular G-quadruplex structures. Mutation and deletion analysis permitted isolation and identification of three overlapping DNA sequences within this element that formed the three individual G-quadruplexes. Each of these was characterized using nondenaturing gel analysis, DMS footprinting, and circular dichroism. The central G-quadruplex, which is the most stable, forms a mixed parallel/antiparallel structure consisting of three tetrads connected by loops of one, seven, and three bases. Three different G-quadruplex-interactive agents were found to further stabilize these structures, with individual selectivity toward one or more of these G-quadruplexes. Collectively, these results suggest that the multiple G-quadruplexes identified in the promoter region of the bcl-2 gene are likely to play a similar role to the G-quadruplexes in the c-myc promoter in that their formation could serve to modulate gene transcription. Last, we demonstrate that the complexity of the G-quadruplexes in the bcl-2 promoter extends beyond the ability to form any one of three separate G-quadruplexes to each having the capacity to form either three or six different loop isomers. These results are discussed in relation to the biological significance of this G-quadruplex-forming element in modulation of bcl-2 gene expression and the inherent complexity of the system where different G-quadruplexes and loop isomers are possible.
Figures







References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

