The three-dimensional structure of complex I from Yarrowia lipolytica: a highly dynamic enzyme
- PMID: 16621601
- PMCID: PMC1764498
- DOI: 10.1016/j.jsb.2006.02.011
The three-dimensional structure of complex I from Yarrowia lipolytica: a highly dynamic enzyme
Abstract
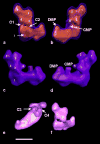
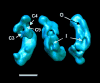
The structure of complex I from Yarrowia lipolytica was determined by three-dimensional electron microscopy. A random conical data set was collected from deep stain embedded particles. More than 14000 image pairs were analyzed. Through extensive classification combined with three-dimensional reconstruction, it was possible for the first time to show a much more detailed substructure of the complex. The peripheral arm is subdivided in at least six domains. The membrane arm shows two major protrusions on its matrix facing side and exhibits a channel like feature on the side facing the cytoplasm. Structures resembling a tether connecting the subunits near the catalytic center with the protrusions of the membrane arm provide a second connection between matrix and membrane domain.
Figures





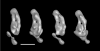
References
-
- Abdrakhmanova A, Zickermann V, Bostina M, Radermacher M, Schagger H, Kerscher S, Brandt U. Subunit composition of mitochondrial complex I from the yeast Yarrowia lipolytica. Biochim Biophys Acta. 2004;1658:148–156. - PubMed
-
- BBA . Special Issue. Biochimica et Biophysica Acta. 1998;1364:89–296. - PubMed
-
- Belogrudov G, Hatefi Y. Catalytic sector of complex I (NADH:ubiquinone oxidoreductase): subunit stoichiometry and substrate-induced conformation changes. Biochemistry. 1994;33:4571–4576. - PubMed
-
- Böttcher B, Scheide D, Hesterberg M, Nagel-Steger L, Friedrich T. A novel, enzymatically active conformation of the Escherichia coli ADH:ubiquinone oxidoreductase (complex I) Journal of Biological Chemistry. 2002;277:17970–17977. - PubMed
-
- Brandt U. Energy converting NADH:quinone oxidoreductase (Complex I) Ann Rev Biochem. 2006;75 in press. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

