Mutational analysis of the Escherichia coli melR gene suggests a two-state concerted model to explain transcriptional activation and repression in the melibiose operon
- PMID: 16621812
- PMCID: PMC1447455
- DOI: 10.1128/JB.188.9.3199-3207.2006
Mutational analysis of the Escherichia coli melR gene suggests a two-state concerted model to explain transcriptional activation and repression in the melibiose operon
Abstract
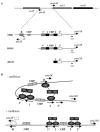
Transcription of the Escherichia coli melAB operon is regulated by the MelR protein, an AraC family member whose activity is modulated by the binding of melibiose. In the absence of melibiose, MelR is unable to activate the melAB promoter but autoregulates its own expression by repressing the melR promoter. Melibiose triggers MelR-dependent activation of the melAB promoter and relieves MelR-dependent repression of the melR promoter. Twenty-nine single amino acid substitutions in MelR that result in partial melibiose-independent activation of the melAB promoter have been identified. Combinations of different substitutions result in almost complete melibiose-independent activation of the melAB promoter. MelR carrying each of the single substitutions is less able to repress the melR promoter, while MelR carrying some combinations of substitutions is completely unable to repress the melR promoter. These results argue that different conformational states of MelR are responsible for activation of the melAB promoter and repression of the melR promoter. Supporting evidence for this is provided by the isolation of substitutions in MelR that block melibiose-dependent activation of the melAB promoter while not changing melibiose-independent repression of the melR promoter. Additional experiments with a bacterial two-hybrid system suggest that interactions between MelR subunits differ according to the two conformational states.
Figures

References
-
- Belyaeva, T. A., J. T. Wade, C. L. Webster, V. J. Howard, M. S. Thomas, E. I. Hyde, and S. J. Busby. 2000. Transcription activation at the Escherichia coli melAB promoter: the role of MelR and the cyclic AMP receptor protein. Mol. Microbiol. 36:211-222. - PubMed
-
- Dunwell, J. M., A. Culham, C. E. Carter, C. R. Sosa-Aguirre, and P. W. Goodenough. 2001. Evolution of functional diversity in the cupin superfamily. Trends Biochem. Sci. 26:740-746. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

