Selenium is involved in regulation of periplasmic hydrogenase gene expression in Desulfovibrio vulgaris Hildenborough
- PMID: 16621815
- PMCID: PMC1447438
- DOI: 10.1128/JB.188.9.3228-3235.2006
Selenium is involved in regulation of periplasmic hydrogenase gene expression in Desulfovibrio vulgaris Hildenborough
Abstract
Desulfovibrio vulgaris Hildenborough is a good model organism to study hydrogen metabolism in sulfate-reducing bacteria. Hydrogen is a key compound for these organisms, since it is one of their major energy sources in natural habitats and also an intermediate in the energy metabolism. The D. vulgaris Hildenborough genome codes for six different hydrogenases, but only three of them, the periplasmic-facing [FeFe], [FeNi]1, and [FeNiSe] hydrogenases, are usually detected. In this work, we studied the synthesis of each of these enzymes in response to different electron donors and acceptors for growth as well as in response to the availability of Ni and Se. The formation of the three hydrogenases was not very strongly affected by the electron donors or acceptors used, but the highest levels were observed after growth with hydrogen as electron donor and lowest with thiosulfate as electron acceptor. The major effect observed was with inclusion of Se in the growth medium, which led to a strong repression of the [FeFe] and [NiFe]1 hydrogenases and a strong increase in the [NiFeSe] hydrogenase that is not detected in the absence of Se. Ni also led to increased formation of the [NiFe]1 hydrogenase, except for growth with H2, where its synthesis is very high even without Ni added to the medium. Growth with H2 results in a strong increase in the soluble forms of the [NiFe]1 and [NiFeSe] hydrogenases. This study is an important contribution to understanding why D. vulgaris Hildenborough has three periplasmic hydrogenases. It supports their similar physiological role in H2 oxidation and reveals that element availability has a strong influence in their relative expression.
Figures


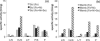

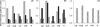

Similar articles
-
Function of periplasmic hydrogenases in the sulfate-reducing bacterium Desulfovibrio vulgaris Hildenborough.J Bacteriol. 2007 Sep;189(17):6159-67. doi: 10.1128/JB.00747-07. Epub 2007 Jun 29. J Bacteriol. 2007. PMID: 17601789 Free PMC article.
-
Characterization of the [NiFeSe] hydrogenase from Desulfovibrio vulgaris Hildenborough.Methods Enzymol. 2018;613:169-201. doi: 10.1016/bs.mie.2018.10.003. Epub 2018 Nov 24. Methods Enzymol. 2018. PMID: 30509465
-
Effects of deletion of genes encoding Fe-only hydrogenase of Desulfovibrio vulgaris Hildenborough on hydrogen and lactate metabolism.J Bacteriol. 2002 Feb;184(3):679-86. doi: 10.1128/JB.184.3.679-686.2002. J Bacteriol. 2002. PMID: 11790737 Free PMC article.
-
Hydrogenases and H2 metabolism in sulfate-reducing bacteria of the Desulfovibrio genus.Adv Microb Physiol. 2019;74:143-189. doi: 10.1016/bs.ampbs.2019.03.001. Epub 2019 Apr 22. Adv Microb Physiol. 2019. PMID: 31126530 Review.
-
The three classes of hydrogenases from sulfate-reducing bacteria of the genus Desulfovibrio.FEMS Microbiol Rev. 1988 Dec;4(4):299-344. doi: 10.1111/j.1574-6968.1988.tb02748.x. FEMS Microbiol Rev. 1988. PMID: 3078655 Review.
Cited by
-
FTIR spectroelectrochemical characterization of the Ni-Fe-Se hydrogenase from Desulfovibrio vulgaris Hildenborough.J Biol Inorg Chem. 2008 Nov;13(8):1315-20. doi: 10.1007/s00775-008-0412-5. Epub 2008 Aug 13. J Biol Inorg Chem. 2008. PMID: 18704522
-
Variation among Desulfovibrio species in electron transfer systems used for syntrophic growth.J Bacteriol. 2013 Mar;195(5):990-1004. doi: 10.1128/JB.01959-12. Epub 2012 Dec 21. J Bacteriol. 2013. PMID: 23264581 Free PMC article.
-
Roles of HynAB and Ech, the only two hydrogenases found in the model sulfate reducer Desulfovibrio gigas.J Bacteriol. 2013 Oct;195(20):4753-60. doi: 10.1128/JB.00411-13. Epub 2013 Aug 23. J Bacteriol. 2013. PMID: 23974026 Free PMC article.
-
Interaction of the active site of the Ni-Fe-Se hydrogenase from Desulfovibrio vulgaris Hildenborough with carbon monoxide and oxygen inhibitors.J Biol Inorg Chem. 2010 Nov;15(8):1285-92. doi: 10.1007/s00775-010-0686-2. Epub 2010 Jul 29. J Biol Inorg Chem. 2010. PMID: 20669037
-
Genetics and molecular biology of the electron flow for sulfate respiration in desulfovibrio.Front Microbiol. 2011 Jun 29;2:135. doi: 10.3389/fmicb.2011.00135. eCollection 2011. Front Microbiol. 2011. PMID: 21747813 Free PMC article.
References
-
- Afting, C., E. Kremmer, C. Brucker, A. Hochheimer, and R. K. Thauer. 2000. Regulation of the synthesis of H-2-forming methylenetetrahydromethanopterin dehydrogenase (Hmd) and of HmdII and HmdIII in Methanothermobacter marburgensis. Arch. Microbiol. 174:225-232. - PubMed
-
- Andrews, S. C., A. K. Robinson, and F. Rodriguez-Quinones. 2003. Bacterial iron homeostasis. FEMS Microbiol. Rev. 27:215-237. - PubMed
-
- Berghofer, Y., K. Agha-Amiri, and A. Klein. 1994. Selenium is involved in the negative regulation of the expression of selenium-free [NiFe] hydrogenases in Methanococcus voltae. Mol. Gen. Genet. 242:369-373. - PubMed
-
- Birringer, M., S. Pilawa, and L. Flohé. 2002. Trends in selenium biochemistry. Nat. Prod. Rep. 19:693-718. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

