A distinct QscR regulon in the Pseudomonas aeruginosa quorum-sensing circuit
- PMID: 16621831
- PMCID: PMC1447466
- DOI: 10.1128/JB.188.9.3365-3370.2006
A distinct QscR regulon in the Pseudomonas aeruginosa quorum-sensing circuit
Abstract
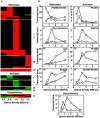
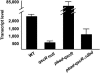
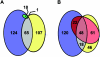
The opportunistic pathogen Pseudomonas aeruginosa possesses two complete acyl-homoserine lactone (acyl-HSL) signaling systems. One system consists of LasI and LasR, which generate a 3-oxododecanoyl-homoserine lactone signal and respond to that signal, respectively. The other system is RhlI and RhlR, which generate butanoyl-homoserine lactone and respond to butanoyl-homoserine lactone, respectively. These quorum-sensing systems control hundreds of genes. There is also an orphan LasR-RhlR homolog, QscR, for which there is no cognate acyl-HSL synthetic enzyme. We previously reported that a qscR mutant is hypervirulent and showed that QscR transiently represses a few quorum-sensing-controlled genes. To better understand the role of QscR in P. aeruginosa gene regulation and to better understand the relationship between QscR, LasR, and RhlR control of gene expression, we used transcription profiling to identify a QscR-dependent regulon. Our analysis revealed that QscR activates some genes and represses others. Some of the repressed genes are not regulated by the LasR-I or RhlR-I systems, while others are. The LasI-generated 3-oxododecanoyl-homoserine lactone serves as a signal molecule for QscR. Thus, QscR appears to be an integral component of the P. aeruginosa quorum-sensing circuitry. QscR uses the LasI-generated acyl-homoserine lactone signal and controls a specific regulon that overlaps with the already overlapping LasR- and RhlR-dependent regulons.
Figures
Comment in
-
The QscR quorum-sensing regulon of Pseudomonas aeruginosa: an orphan claims its identity.J Bacteriol. 2006 May;188(9):3169-71. doi: 10.1128/JB.188.9.3169-3171.2006. J Bacteriol. 2006. PMID: 16621807 Free PMC article. No abstract available.
References
-
- Baldi, P., and A. D. Long. 2001. A Bayesian framework for the analysis of microarray expression data: regularized t-test and statistical inferences of gene changes. Bioinformatics 17:509-519. - PubMed
-
- Chai, Y., J. Zhu, and S. C. Winans. 2001. TrlR, a defective TraR-like protein of Agrobacterium tumefaciens, blocks TraR function in vitro by forming inactive TrlR:TraR dimers. Mol. Microbiol. 40:414-421. - PubMed
-
- Davies, D. G., M. R. Parsek, J. P. Pearson, B. H. Iglewski, J. W. Costerton, and E. P. Greenberg. 1998. The involvement of cell-to-cell signals in the development of a bacterial biofilm. Science 280:295-298. - PubMed
-
- Fuqua, C., and E. P. Greenberg. 2002. Listening in on bacteria: acyl-homoserine lactone signalling. Nat. Rev. Mol. Cell Biol. 3:685-695. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

