Transcriptional regulatory networks downstream of TAL1/SCL in T-cell acute lymphoblastic leukemia
- PMID: 16621969
- PMCID: PMC1895859
- DOI: 10.1182/blood-2005-08-3482
Transcriptional regulatory networks downstream of TAL1/SCL in T-cell acute lymphoblastic leukemia
Abstract
Aberrant expression of 1 or more transcription factor oncogenes is a critical component of the molecular pathogenesis of human T-cell acute lymphoblastic leukemia (T-ALL); however, oncogenic transcriptional programs downstream of T-ALL oncogenes are mostly unknown. TAL1/SCL is a basic helix-loop-helix (bHLH) transcription factor oncogene aberrantly expressed in 60% of human T-ALLs. We used chromatin immunoprecipitation (ChIP) on chip to identify 71 direct transcriptional targets of TAL1/SCL. Promoters occupied by TAL1 were also frequently bound by the class I bHLH proteins E2A and HEB, suggesting that TAL1/E2A as well as TAL1/HEB heterodimers play a role in transformation of T-cell precursors. Using RNA interference, we demonstrated that TAL1 is required for the maintenance of the leukemic phenotype in Jurkat cells and showed that TAL1 binding can be associated with either repression or activation of genes whose promoters occupied by TAL1, E2A, and HEB. In addition, oligonucleotide microarray analysis of RNA from 47 primary T-ALL samples showed specific expression signatures involving TAL1 targets in TAL1-expressing compared with -nonexpressing human T-ALLs. Our results indicate that TAL1 may act as a bifunctional transcriptional regulator (activator and repressor) at the top of a complex regulatory network that disrupts normal T-cell homeostasis and contributes to leukemogenesis.
Figures


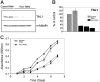
 ) or the TAL1 shRNA (▪) reveals 70% to 90% knockdown of TAL1 at the RNA level in TAL1 knockdown cells. Error bars indicate standard deviations. (C) TAL1 knockdown affects the growth rate of Jurkat cells. Cells (5 × 104) from clones stably expressing a control or TAL1 shRNAs were plated in RPMI with 5% FBS, and the cell numbers were measured daily using an MTT-based assay.
) or the TAL1 shRNA (▪) reveals 70% to 90% knockdown of TAL1 at the RNA level in TAL1 knockdown cells. Error bars indicate standard deviations. (C) TAL1 knockdown affects the growth rate of Jurkat cells. Cells (5 × 104) from clones stably expressing a control or TAL1 shRNAs were plated in RPMI with 5% FBS, and the cell numbers were measured daily using an MTT-based assay.

References
-
- Porcher C, Swat W, Rockwell K, Fujiwara Y, Alt FW, Orkin SH. The T cell leukemia oncoprotein SCL/tal-1 is essential for development of all hematopoietic lineages. Cell. 1996;86: 47-57. - PubMed
-
- Robb L, Begley CG. The SCL/TAL1 gene: roles in normal and malignant haematopoiesis. Bioessays. 1997;19: 607-613. - PubMed
-
- Ferrando AA, Neuberg DS, Staunton J, et al. Gene expression signatures define novel oncogenic pathways in T cell acute lymphoblastic leukemia. Cancer Cell. 2002;1: 75-87. - PubMed
-
- Ferrando AA, Neuberg DS, Dodge RK, et al. Prognostic importance of TLX1 (HOX11) oncogene expression in adults with T-cell acute lymphoblastic leukaemia. Lancet. 2004;363: 535-536. - PubMed
-
- Ferrando AA, Herblot S, Palomero T, et al. Biallelic transcriptional activation of oncogenic transcription factors in T-cell acute lymphoblastic leukemia. Blood. 2004;103: 1909-1911. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

