The most polymorphic residue on Plasmodium falciparum apical membrane antigen 1 determines binding of an invasion-inhibitory antibody
- PMID: 16622199
- PMCID: PMC1459722
- DOI: 10.1128/IAI.74.5.2628-2636.2006
The most polymorphic residue on Plasmodium falciparum apical membrane antigen 1 determines binding of an invasion-inhibitory antibody
Abstract
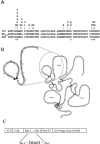
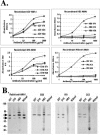
Apical membrane antigen 1 (AMA1) is currently one of the leading malarial vaccine candidates. Anti-AMA1 antibodies can inhibit the invasion of erythrocytes by Plasmodium merozoites and prevent the multiplication of blood-stage parasites. Here we describe an anti-AMA1 monoclonal antibody (MAb 1F9) that inhibits the invasion of Plasmodium falciparum parasites in vitro. We show that both reactivity of MAb 1F9 with AMA1 and MAb 1F9-mediated invasion inhibition were strain specific. Site-directed mutagenesis of a fragment of AMA1 displayed on M13 bacteriophage identified a single polymorphic residue in domain I of AMA1 that is critical for MAb 1F9 binding. The identities of all other polymorphic residues investigated in this domain had little effect on the binding of the antibody. Examination of the P. falciparum AMA1 crystal structure localized this residue to a surface-exposed alpha-helix at the apex of the polypeptide. This description of a polymorphic inhibitory epitope on AMA1 adds supporting evidence to the hypothesis that immune pressure is responsible for the polymorphisms seen in this molecule.
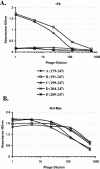
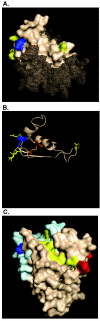
Figures
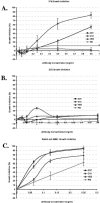
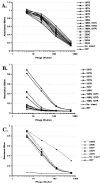
References
-
- Anders, R. F., P. E. Crewther, S. Edwards, M. Margetts, M. L. Matthew, B. Pollock, and D. Pye. 1998. Immunisation with recombinant AMA-1 protects mice against infection with Plasmodium chabaudi. Vaccine 16:240-247. - PubMed
-
- Anders, R. F., D. J. McColl, and R. L. Coppel. 1993. Molecular variation in Plasmodium falciparum: polymorphic antigens of asexual erythrocytic stages. Acta Trop. 53:239-253. - PubMed
-
- Basco, L. K., F. Marquet, M. M. Makler, and J. Le Bras. 1995. Plasmodium falciparum and Plasmodium vivax: lactate dehydrogenase activity and its application for in vitro drug susceptibility assay. Exp. Parasitol. 80:260-271. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

