How dietary arachidonic- and docosahexaenoic- acid rich oils differentially affect the murine hepatic transcriptome
- PMID: 16623957
- PMCID: PMC1479345
- DOI: 10.1186/1476-511X-5-10
How dietary arachidonic- and docosahexaenoic- acid rich oils differentially affect the murine hepatic transcriptome
Abstract
Introduction: Herein, we expand our previous work on the effects of long chain polyunsaturated fatty acids (LC-PUFA) on the murine hepatic transcriptome using novel statistical and bioinformatic approaches for evaluating microarray data. The analyses focuses on key differences in the transcriptomic response that will influence metabolism following consumption of FUNG (rich in 20:4n6), FISH (rich in 20:5n3, 22:5n3, and 22:6n3) and COMB, the combination of the two.
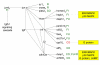
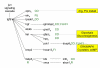
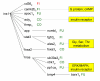
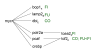
Results: Using a variance-stabilized F-statistic, 371 probe sets (out of 13 K probe sets in the Affymetrix Mu11K chip set) were changed by dietary treatment (P < 0.001). Relative to other groups, COMB had unique affects on murine hepatic transcripts involved in cytoskeletal and carbohydrate metabolism; whereas FUNG affected amino acid metabolism via CTNB1 signaling. All three diets affected transcripts linked to apoptosis and cell proliferation, with evidence FISH may have increased apoptosis and decreased cell proliferation via various transcription factors, kinases, and phosphatases. The three diets affected lipid transport, lipoprotein metabolism, and bile acid metabolism through diverse pathways. Relative to other groups, FISH activated cyps that form hydroxylated fatty acids known to affect vascular tone and ion channel activity. FA synthesis and delta 9 desaturation were down regulated by COMB relative to other groups, implying that a FA mixture of 20:4n6, 20:5n3, and 22:6n3 is most effective at down regulating synthesis, via INS1, SREBP, PPAR alpha, and TNF signaling. Heme synthesis and the utilization of heme for hemoglobin production were likely affected by FUNG and FISH. Finally, relative to other groups, FISH increased numerous transcripts linked to combating oxidative such as peroxidases, an aldehyde dehydrogenase, and heat shock proteins, consistent with the major LC-PUFA in FISH (20:5n3, 22:5n3, 22:6n3) being more oxidizable than the major fatty acids in FUNG (20:4n6).
Conclusion: Distinct transcriptomic, signaling cascades, and predicted affects on murine liver metabolism have been elucidated for 20:4n6-rich dietary oils, 22:6n3-rich oils, and a surprisingly distinct set of genes were affected by the combination of the two. Our results emphasize that the balance of dietary n6 and n3 LC-PUFA provided for infants and in nutritional and neutraceutical applications could have profoundly different affects on metabolism and cell signaling, beyond that previously recognized.
Figures



References
-
- Lapillonne A, Clarke SD, Heird WC. Polyunsaturated fatty acids and gene expression. Curr Opin Clin Nutr Metab Care. 2004;7:151–156. - PubMed
-
- Berger A, Roberts MA. Unraveling lipid metabolism with microarrays. NY, Marcel Dekker (Taylor & Francis, CRC Press); 2005. p. 445.
-
- Ide T, Kobayashi H, Ashakumary L, Rouyer IA, Takahashi Y, Aoyama T, Hashimoto T, Mizugaki M. Comparative effects of perilla and fish oils on the activity and gene expression of fatty acid oxidation enzymes in rat liver. Biochim Biophys Acta. 2000;1485:23–35. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

