Site-specific proteolysis of the transcriptional coactivator HCF-1 can regulate its interaction with protein cofactors
- PMID: 16624878
- PMCID: PMC1440766
- DOI: 10.1073/pnas.0602109103
Site-specific proteolysis of the transcriptional coactivator HCF-1 can regulate its interaction with protein cofactors
Abstract
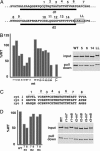
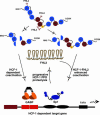
Limited proteolytic processing is an important transcriptional regulatory mechanism. In various contexts, proteolysis controls the cytoplasmic-to-nuclear transport of important transcription factors or removes domains to produce factors with altered activities. The transcriptional coactivator host cell factor-1 (HCF-1) is proteolytically processed within a unique domain consisting of 20-aa reiterations. Site-specific cleavage within one or more repeats generates a family of amino- and carboxyl-terminal subunits that remain tightly associated. However, the consequences of HCF-1 processing have been undefined. In this study, it was determined that the HCF-1-processing domain interacts with several proteins including the transcriptional coactivator/corepressor four-and-a-half LIM domain-2 (FHL2). Analysis of this interaction has uncovered specificity with both sequence and context determinants within the reiterations of this processing domain. In cells, FHL2 interacts exclusively with the nonprocessed coactivator and costimulates transcription of an HCF-1-dependent target gene. The functional interaction of HCF-1 with FHL2 supports a model in which site-specific proteolysis regulates the interaction of HCF-1 with protein partners and thus can modulate the activity of this coactivator. This paradigm expands the biological significance of limited proteolytic processing as a regulatory mechanism in gene transcription.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

