Fission yeast Num1p is a cortical factor anchoring dynein and is essential for the horse-tail nuclear movement during meiotic prophase
- PMID: 16624923
- PMCID: PMC1526665
- DOI: 10.1534/genetics.105.050062
Fission yeast Num1p is a cortical factor anchoring dynein and is essential for the horse-tail nuclear movement during meiotic prophase
Abstract
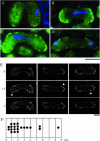
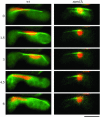
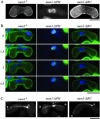
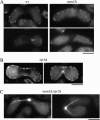
During meiotic prophase in the fission yeast Schizosaccharomyces pombe, the nucleus oscillates between the two ends of a cell. This oscillatory nuclear movement is important to promote accurate pairing of homologous chromosomes and requires cytoplasmic dynein. Dynein accumulates at the points where microtubule plus ends contact the cell cortex and generate a force to drive nuclear oscillation. However, it remains poorly understood how dynein associates with the cell cortex. Here we show that S. pombe Num1p functions as a cortical-anchoring factor for dynein. Num1p is expressed in a meiosis-specific manner and localized to the cell cortex through its C-terminal PH domain. The num1 deletion mutant shows microtubule dynamics comparable to that in the wild type. However, it lacks cortical accumulation of dynein and is defective in the nuclear oscillation as is the case for the dynein mutant. We also show that Num1p can recruit dynein independently of the CLIP-170 homolog Tip1p.
Figures



References
-
- Bahler, J., J. Q. Wu, M. S. Longtine, N. G. Shah, A. McKenzie III et al., 1998. Heterologous modules for efficient and versatile PCR-based gene targeting in Schizosaccharomyces pombe. Yeast 14: 943–951. - PubMed
-
- Basi, G., E. Schmid and K. Maundrell, 1993. TATA box mutations in the Schizosaccharomyces pombe nmt1 promotor affect transcription efficiency but not the transcription start point or thiamine repressibility. Gene 123: 131–136. - PubMed
-
- Bloom, K., 2001. Nuclear migration: cortical anchors for cytoplasmic dynein. Curr. Biol. 11: R326–R329. - PubMed
-
- Brunner, D., and P. Nurse, 2000. CLIP170-like tip1p spatially organizes microtubular dynamics in fission yeast. Cell 102: 695–704. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

