Classification and risk stratification of invasive breast carcinomas using a real-time quantitative RT-PCR assay
- PMID: 16626501
- PMCID: PMC1557722
- DOI: 10.1186/bcr1399
Classification and risk stratification of invasive breast carcinomas using a real-time quantitative RT-PCR assay
Abstract
Introduction: Predicting the clinical course of breast cancer is often difficult because it is a diverse disease comprised of many biological subtypes. Gene expression profiling by microarray analysis has identified breast cancer signatures that are important for prognosis and treatment. In the current article, we use microarray analysis and a real-time quantitative reverse-transcription (qRT)-PCR assay to risk-stratify breast cancers based on biological 'intrinsic' subtypes and proliferation.
Methods: Gene sets were selected from microarray data to assess proliferation and to classify breast cancers into four different molecular subtypes, designated Luminal, Normal-like, HER2+/ER-, and Basal-like. One-hundred and twenty-three breast samples (117 invasive carcinomas, one fibroadenoma and five normal tissues) and three breast cancer cell lines were prospectively analyzed using a microarray (Agilent) and a qRT-PCR assay comprised of 53 genes. Biological subtypes were assigned from the microarray and qRT-PCR data by hierarchical clustering. A proliferation signature was used as a single meta-gene (log2 average of 14 genes) to predict outcome within the context of estrogen receptor status and biological 'intrinsic' subtype.
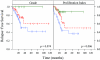
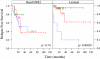
Results: We found that the qRT-PCR assay could determine the intrinsic subtype (93% concordance with microarray-based assignments) and that the intrinsic subtypes were predictive of outcome. The proliferation meta-gene provided additional prognostic information for patients with the Luminal subtype (P = 0.0012), and for patients with estrogen receptor-positive tumors (P = 3.4 x 10-6). High proliferation in the Luminal subtype conferred a 19-fold relative risk of relapse (confidence interval = 95%) compared with Luminal tumors with low proliferation.
Conclusion: A real-time qRT-PCR assay can recapitulate microarray classifications of breast cancer and can risk-stratify patients using the intrinsic subtype and proliferation. The proliferation meta-gene offers an objective and quantitative measurement for grade and adds significant prognostic information to the biological subtypes.
Figures



References
-
- American Joint Committee on Cancer http://www.cancerstaging.org/cstage/index.html
-
- Elston CW, Ellis IO. Pathological prognostic factors in breast cancer. I. The value of histological grade in breast cancer: experience from a large study with long-term follow-up. Histopathology. 1991;19:403–410. - PubMed
-
- Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS, et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci USA. 2001;98:10869–10874. doi: 10.1073/pnas.191367098. - DOI - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

