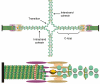
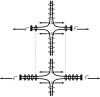
The path of DNA in the kinetochore
- PMID: 16631569
- PMCID: PMC2112771
- DOI: 10.1016/j.cub.2006.03.054
The path of DNA in the kinetochore
Figures
References
-
- McAinsh AD, Tytell JD, Sorger PK. Structure, function, and regulation of budding yeast kinetochores. Annu. Rev Cell Dev. Biol. 2003;19:519–539. - PubMed
-
- Blat Y, Kleckner N. Cohesins bind to preferential sites along yeast chromosome III, with differential regulation along arms versus the centric region. Cell. 1999;98:249–259. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

