Functional analysis of the validamycin biosynthetic gene cluster and engineered production of validoxylamine A
- PMID: 16632251
- PMCID: PMC1474575
- DOI: 10.1016/j.chembiol.2006.02.002
Functional analysis of the validamycin biosynthetic gene cluster and engineered production of validoxylamine A
Abstract
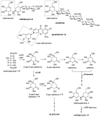
A 45 kb DNA sequencing analysis from Streptomyces hygroscopicus 5008 involved in validamycin A (VAL-A) biosynthesis revealed 16 structural genes, 2 regulatory genes, 5 genes related transport, transposition/integration or tellurium resistance; another 4 genes had no obvious identity. The VAL-A biosynthetic pathway was proposed, with assignment of the required genetic functions confined to the sequenced region. A cluster of eight reassembled genes was found to support VAL-A synthesis in a heterologous host, S. lividans 1326. In vivo inactivation of the putative glycosyltransferase gene (valG) abolished the final attachment of glucose for VAL production and resulted in accumulation of the VAL-A precursor, validoxylamine, while the normal production of VAL-A could be restored by complementation with valG. The role of valG in the glycosylation of validoxylamine to VAL-A was demonstrated in vitro by enzymatic assay.
Figures





References
-
- Iwasa T, Yamamoto H, Shibata M. Studies on validamycins, new antibiotics. I. Streptomyces hygroscopicus var. limoneus nov. var., validamycin-producing organism. Jpn J Antibiot. 1970;23:595–602. - PubMed
-
- Iwasa T, Higashide E, Yamamoto H, Shibata M. Studies on validamycins, new antibiotics. II. Production and biological properties of validamycins A and B. J Antibiot (Tokyo) 1971;24:107–113. - PubMed
-
- Asano N, Takeuchi M, Kameda Y, Matsui K, Kono Y. Trehalase inhibitors, validoxylamine A and related compounds as insecticides. J Antibiot (Tokyo) 1990;43:722–726. - PubMed
-
- Kameda Y, Asano N, Yamaguchi T, Matsui K. Validoxylamines as trehalase inhibitors. J Antibiot (Tokyo) 1987;40:563–565. - PubMed
-
- Asano N, Tanaka K, Kameda Y, Matsui K. All eight possible mono-beta-D-glucosides of validoxylamine A. II. Biological activities. J Antibiot (Tokyo) 1991;44:1417–1421. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

