Weak selection and recent mutational changes influence polymorphic synonymous mutations in humans
- PMID: 16632609
- PMCID: PMC1458998
- DOI: 10.1073/pnas.0510638103
Weak selection and recent mutational changes influence polymorphic synonymous mutations in humans
Abstract
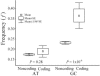
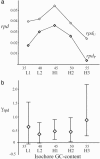
Recent large-scale genomic and evolutionary studies have revealed the small but detectable signature of weak selection on synonymous mutations during mammalian evolution, likely acting at the level of translational efficacy (i.e., translational selection). To investigate whether weak selection, and translational selection in particular, plays any role in shaping the fate of synonymous mutations that are present today in human populations, we studied genetic variation at the polymorphic level and patterns of evolution in the human lineage after human-chimpanzee separation. We find evidence that neutral mechanisms are influencing the frequency of polymorphic mutations in humans. Our results suggest a recent increase in mutational tendencies toward AT, observed in all isochores, that is responsible for AT mutations segregating at lower frequencies than GC mutations. In all, however, changes in mutational tendencies and other neutral scenarios are not sufficient to explain a difference between synonymous and noncoding mutations or a difference between synonymous mutations potentially advantageous or deleterious under a translational selection model. Furthermore, several estimates of selection intensity on synonymous mutations all suggest a detectable influence of weak selection acting at the level of translational selection. Thus, random genetic drift, recent changes in mutational tendencies, and weak selection influence the fate of synonymous mutations that are present today as polymorphisms. All of these features, neutral and selective, should be taken into account in evolutionary analyses that often assume constancy of mutational tendencies and complete neutrality of synonymous mutations.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
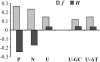


References
-
- Bernardi G., Olofsson B., Filipski J., Zerial M., Salinas J., Cuny G., Meunier-Rotival M., Rodier F. Science. 1985;228:953–958. - PubMed
-
- Eyre-Walker A., Hurst L. D. Nat. Rev. Genet. 2001;2:549–555. - PubMed
-
- Ikemura T. Mol. Biol. Evol. 1985;2:13–34. - PubMed
-
- Sharp P. M., Averof M., Lloyd A. T., Matassi G., Peden J. F. Philos. Trans. R. Soc. London B. 1995;349:241–247. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

