Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy
- PMID: 16632643
- PMCID: PMC1475499
- DOI: 10.1105/tpc.106.041665
Comparative genomics of Brassica oleracea and Arabidopsis thaliana reveal gene loss, fragmentation, and dispersal after polyploidy
Abstract
We sequenced 2.2 Mb representing triplicated genome segments of Brassica oleracea, which are each paralogous with one another and homologous with a segmentally duplicated region of the Arabidopsis thaliana genome. Sequence annotation identified 177 conserved collinear genes in the B. oleracea genome segments. Analysis of synonymous base substitution rates indicated that the triplicated Brassica genome segments diverged from a common ancestor soon after divergence of the Arabidopsis and Brassica lineages. This conclusion was corroborated by phylogenetic analysis of protein families. Using A. thaliana as an outgroup, 35% of the genes inferred to be present when genome triplication occurred in the Brassica lineage have been lost, most likely via a deletion mechanism, in an interspersed pattern. Genes encoding proteins involved in signal transduction or transcription were not found to be significantly more extensively retained than those encoding proteins classified with other functions, but putative proteins predicted in the A. thaliana genome were underrepresented in B. oleracea. We identified one example of gene loss from the Arabidopsis lineage. We found evidence for the frequent insertion of gene fragments of nuclear genomic origin and identified four apparently intact genes in noncollinear positions in the B. oleracea and A. thaliana genomes.
Figures
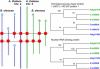

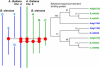
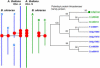
References
-
- Arabidopsis Genome Initiative (2000). Analysis of the genome of the flowering plant Arabidopsis thaliana. Nature 408 796–815. - PubMed
-
- Arumuganthan, K., and Earle, E.D. (1991). Nuclear DNA content of some important plant species. Plant Mol. Biol. Rep. 9 208–218.
-
- Babula, D., Kaczmarek, M., Barakat, A., Delseney, M., Quiros, C.F., and Sadowski, J. (2003). Chromosomal mapping of Brassica oleracea based on ESTs from Arabidopsis thaliana: Complexity of the comparative map. Mol. Genet. Genomics 268 656–665. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

