Proton movement and photointermediate kinetics in rhodopsin mutants
- PMID: 16634624
- PMCID: PMC2527178
- DOI: 10.1021/bi0525775
Proton movement and photointermediate kinetics in rhodopsin mutants
Abstract
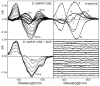
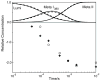
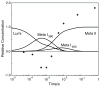
The role of ionizable amino acid side chains in the bovine rhodopsin activation mechanism was studied in mutants E134Q, E134R/R135E, H211F, and E122Q. All mutants exhibited bathorhodopsin stability on the 30 ns to 1 micros time scale similar to that of the wild type. Lumirhodopsin decay was also similar to that of the wild type except for the H211F mutant where early decay (20 micros) to a second form of lumirhodopsin was seen, followed by formation of an extremely long-lived Meta I(480) product (34 ms), an intermediate which forms to a much reduced extent, if at all, in dodecyl maltoside suspensions of wild-type rhodopsin. A smaller amount of a similar long-lived Meta I(480) product was seen after photolysis of E122Q, but E134Q and E134R/R135Q displayed kinetics much more similar to those of the wild type under these conditions (i.e., no Meta I(480) product). These results support the idea that specific interaction of His211 and Glu122 plays a significant role in deprotonation of the retinylidene Schiff base and receptor activation. Proton uptake measurements using bromcresol purple showed that E122Q was qualitatively similar to wild-type rhodopsin, with at least one proton being released during lumirhodopsin decay per Meta I(380) intermediate formed, followed by uptake of at least two protons per rhodopsin bleached on a time scale of tens of milliseconds. Different results were obtained for H211F, E134Q, and E134R/R135E, which all released approximately two protons per rhodopsin bleached. These results show that several ionizable groups besides the Schiff base imine are affected by the structural changes involved in rhodopsin activation. At least two proton uptake groups and probably at least one proton release group in addition to the Schiff base are present in rhodopsin.
Figures





Similar articles
-
Time-resolved photointermediate changes in rhodopsin glutamic acid 181 mutants.Biochemistry. 2004 Oct 5;43(39):12614-21. doi: 10.1021/bi049581l. Biochemistry. 2004. PMID: 15449951
-
pH dependence of photolysis intermediates in the photoactivation of rhodopsin mutant E113Q.Biochemistry. 2000 Jan 25;39(3):599-606. doi: 10.1021/bi991860z. Biochemistry. 2000. PMID: 10642185
-
Identification of glutamic acid 113 as the Schiff base proton acceptor in the metarhodopsin II photointermediate of rhodopsin.Biochemistry. 1994 Sep 13;33(36):10878-82. doi: 10.1021/bi00202a005. Biochemistry. 1994. PMID: 7916209
-
Spectroscopic evidence for interaction between transmembrane helices 3 and 5 in rhodopsin.Biochemistry. 1998 May 19;37(20):7630-9. doi: 10.1021/bi9801560. Biochemistry. 1998. PMID: 9585578
-
Coupling of protonation switches during rhodopsin activation.Photochem Photobiol. 2007 Mar-Apr;83(2):286-92. doi: 10.1562/2006-06-19-IR-937. Photochem Photobiol. 2007. PMID: 17576345 Review.
Cited by
-
A minimal ligand binding pocket within a network of correlated mutations identified by multiple sequence and structural analysis of G protein coupled receptors.BMC Biophys. 2012 Jun 29;5:13. doi: 10.1186/2046-1682-5-13. BMC Biophys. 2012. PMID: 22748306 Free PMC article.
-
Chlorophyll-Derivative Modulation of Rhodopsin Signaling Properties through Evolutionarily Conserved Interaction Pathways.Front Mol Biosci. 2017 Dec 12;4:85. doi: 10.3389/fmolb.2017.00085. eCollection 2017. Front Mol Biosci. 2017. PMID: 29312953 Free PMC article.
-
Amino acid conservation and interactions in rhodopsin: probing receptor activation by NMR spectroscopy.Biochim Biophys Acta. 2014 May;1837(5):683-93. doi: 10.1016/j.bbabio.2013.10.007. Epub 2013 Oct 29. Biochim Biophys Acta. 2014. PMID: 24183693 Free PMC article. Review.
-
Rhodopsin in nanodiscs has native membrane-like photointermediates.Biochemistry. 2011 Jun 7;50(22):5086-91. doi: 10.1021/bi200391a. Epub 2011 May 13. Biochemistry. 2011. PMID: 21539361 Free PMC article.
-
Light activation of rhodopsin: insights from molecular dynamics simulations guided by solid-state NMR distance restraints.J Mol Biol. 2010 Feb 26;396(3):510-27. doi: 10.1016/j.jmb.2009.12.003. Epub 2009 Dec 11. J Mol Biol. 2010. PMID: 20004206 Free PMC article.
References
-
- Vogel R, Siebert F. Fourier transform IR spectroscopy study for new insights into molecular properties and activation mechanisms of visual pigment rhodopsin. Biopolymers. 2003;72:133–148. - PubMed
-
- Okada T, Sugihara M, Bondar AN, Elstner M, Entel P, Buss V. The retinal conformation and its environment in rhodopsin in light of a new 2.2 angstrom crystal structure. J Mol Biol. 2004;342:571–583. - PubMed
-
- Li J, Edwards PC, Burghammer M, Villa C, Schertler GFX. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–1438. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

