Dual targeting of a single tRNA(Trp) requires two different tryptophanyl-tRNA synthetases in Trypanosoma brucei
- PMID: 16636268
- PMCID: PMC1458982
- DOI: 10.1073/pnas.0602362103
Dual targeting of a single tRNA(Trp) requires two different tryptophanyl-tRNA synthetases in Trypanosoma brucei
Abstract
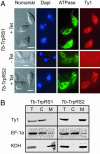
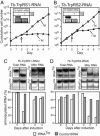
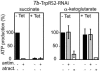
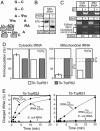
The mitochondrion of Trypanosoma brucei does not encode any tRNAs. This deficiency is compensated for by the import of a small fraction of nearly all of its cytosolic tRNAs. Most trypanosomal aminoacyl-tRNA synthetases are encoded by single-copy genes, suggesting the use of the same enzyme in the cytosol and mitochondrion. However, the T. brucei genome contains two distinct genes for eukaryotic tryptophanyl-tRNA synthetase (TrpRS). RNA interference analysis established that both TrpRS1 and TrpRS2 are essential for growth and required for cytosolic and mitochondrial tryptophanyl-tRNA formation, respectively. Decoding the mitochondrial tryptophan codon UGA requires mitochondria-specific C-->U RNA editing in the anticodon of the imported tRNA(Trp). In vitro charging assays with recombinant TrpRS enzymes demonstrated that the edited anticodon and the mitochondria-specific thiolation of U33 in the imported tRNA(Trp) act as antideterminants for the cytosolic TrpRS1. The existence of two TrpRS enzymes, therefore, can be explained by the need for a mitochondrial synthetase with extended substrate specificity to achieve aminoacylation of the imported thiolated and edited tRNA(Trp). Thus, the notion that, in an organism, all nuclear-encoded tRNAs assigned to a given amino acid are charged by a single aminoacyl-tRNA synthetase, is not universally valid.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures

References
-
- Bullerwell C. E., Gray M. W. Curr. Opin. Microbiol. 2004;7:528–534. - PubMed
-
- Small I., Wintz H., Akashi K., Mireau H. Plant Mol. Biol. 1998;38:265–277. - PubMed
-
- Knight R. D., Freeland S. J., Landweber L. F. Nat. Rev. Genet. 2001;2:49–58. - PubMed
-
- Jorgensen R., Sogaard T. M. M., Rossing A. B., Martensen P. M., Justesen J. J. Biol. Chem. 2000;275:16820–16826. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

