Characterization of hARD2, a processed hARD1 gene duplicate, encoding a human protein N-alpha-acetyltransferase
- PMID: 16638120
- PMCID: PMC1475586
- DOI: 10.1186/1471-2091-7-13
Characterization of hARD2, a processed hARD1 gene duplicate, encoding a human protein N-alpha-acetyltransferase
Abstract
Background: Protein acetylation is increasingly recognized as an important mechanism regulating a variety of cellular functions. Several human protein acetyltransferases have been characterized, most of them catalyzing epsilon-acetylation of histones and transcription factors. We recently described the human protein acetyltransferase hARD1 (human Arrest Defective 1). hARD1 interacts with NATH (N-Acetyl Transferase Human) forming a complex expressing protein N-terminal alpha-acetylation activity.
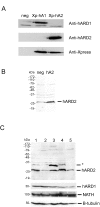
Results: We here describe a human protein, hARD2, with 81 % sequence identity to hARD1. The gene encoding hARD2 most likely originates from a eutherian mammal specific retrotransposition event. hARD2 mRNA and protein are expressed in several human cell lines. Immunoprecipitation experiments show that hARD2 protein potentially interacts with NATH, suggesting that hARD2-NATH complexes may be responsible for protein N-alpha-acetylation in human cells. In NB4 cells undergoing retinoic acid mediated differentiation, the level of endogenous hARD1 and NATH protein decreases while the level of hARD2 protein is stable.
Conclusion: A human protein N-alpha-acetyltransferase is herein described. ARD2 potentially complements the functions of ARD1, adding more flexibility and complexity to protein N-alpha-acetylation in human cells as compared to lower organisms which only have one ARD.
Figures






Similar articles
-
Identification and characterization of the human ARD1-NATH protein acetyltransferase complex.Biochem J. 2005 Mar 15;386(Pt 3):433-43. doi: 10.1042/BJ20041071. Biochem J. 2005. PMID: 15496142 Free PMC article.
-
Induction of apoptosis in human cells by RNAi-mediated knockdown of hARD1 and NATH, components of the protein N-alpha-acetyltransferase complex.Oncogene. 2006 Jul 20;25(31):4350-60. doi: 10.1038/sj.onc.1209469. Epub 2006 Mar 6. Oncogene. 2006. PMID: 16518407
-
A novel human NatA Nalpha-terminal acetyltransferase complex: hNaa16p-hNaa10p (hNat2-hArd1).BMC Biochem. 2009 May 29;10:15. doi: 10.1186/1471-2091-10-15. BMC Biochem. 2009. PMID: 19480662 Free PMC article.
-
The protein acetyltransferase ARD1: a novel cancer drug target?Curr Cancer Drug Targets. 2008 Nov;8(7):545-53. doi: 10.2174/156800908786241113. Curr Cancer Drug Targets. 2008. PMID: 18991565 Review.
-
Versatility of ARD1/NAA10-mediated protein lysine acetylation.Exp Mol Med. 2018 Jul 27;50(7):1-13. doi: 10.1038/s12276-018-0100-7. Exp Mol Med. 2018. PMID: 30054464 Free PMC article. Review.
Cited by
-
The chaperone-like protein HYPK acts together with NatA in cotranslational N-terminal acetylation and prevention of Huntingtin aggregation.Mol Cell Biol. 2010 Apr;30(8):1898-909. doi: 10.1128/MCB.01199-09. Epub 2010 Feb 12. Mol Cell Biol. 2010. PMID: 20154145 Free PMC article.
-
Proteomics analyses reveal the evolutionary conservation and divergence of N-terminal acetyltransferases from yeast and humans.Proc Natl Acad Sci U S A. 2009 May 19;106(20):8157-62. doi: 10.1073/pnas.0901931106. Epub 2009 May 6. Proc Natl Acad Sci U S A. 2009. PMID: 19420222 Free PMC article.
-
NAA10 mutation causing a novel intellectual disability syndrome with Long QT due to N-terminal acetyltransferase impairment.Sci Rep. 2015 Nov 2;5:16022. doi: 10.1038/srep16022. Sci Rep. 2015. PMID: 26522270 Free PMC article.
-
N-α-acetyltransferase 10 (NAA10) in development: the role of NAA10.Exp Mol Med. 2018 Jul 27;50(7):1-11. doi: 10.1038/s12276-018-0105-2. Exp Mol Med. 2018. PMID: 30054454 Free PMC article. Review.
-
Evaluating possible maternal effect lethality and genetic background effects in Naa10 knockout mice.bioRxiv [Preprint]. 2024 Jan 10:2023.04.27.538618. doi: 10.1101/2023.04.27.538618. bioRxiv. 2024. Update in: PLoS One. 2024 May 7;19(5):e0301328. doi: 10.1371/journal.pone.0301328. PMID: 37163119 Free PMC article. Updated. Preprint.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

