Differential expression of selected histone modifier genes in human solid cancers
- PMID: 16638127
- PMCID: PMC1475574
- DOI: 10.1186/1471-2164-7-90
Differential expression of selected histone modifier genes in human solid cancers
Abstract
Background: Post-translational modification of histones resulting in chromatin remodelling plays a key role in the regulation of gene expression. Here we report characteristic patterns of expression of 12 members of 3 classes of chromatin modifier genes in 6 different cancer types: histone acetyltransferases (HATs)- EP300, CREBBP, and PCAF; histone deacetylases (HDACs)- HDAC1, HDAC2, HDAC4, HDAC5, HDAC7A, and SIRT1; and histone methyltransferases (HMTs)- SUV39H1and SUV39H2. Expression of each gene in 225 samples (135 primary tumours, 47 cancer cell lines, and 43 normal tissues) was analysedby QRT-PCR, normalized with 8 housekeeping genes, and given as a ratio by comparison with a universal reference RNA.
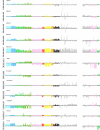
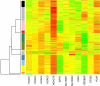
Results: This involved a total of 13,000 PCR assays allowing for rigorous analysis by fitting a linear regression model to the data. Mutation analysis of HDAC1, HDAC2, SUV39H1, and SUV39H2 revealed only two out of 181 cancer samples (both cell lines) with significant coding-sequence alterations. Supervised analysis and Independent Component Analysis showed that expression of many of these genes was able to discriminate tumour samples from their normal counterparts. Clustering based on the normalized expression ratios of the 12 genes also showed that most samples were grouped according to tissue type. Using a linear discriminant classifier and internal cross-validation revealed that with as few as 5 of the 12 genes, SIRT1, CREBBP, HDAC7A, HDAC5 and PCAF, most samples were correctly assigned.
Conclusion: The expression patterns of HATs, HDACs, and HMTs suggest these genes are important in neoplastic transformation and have characteristic patterns of expression depending on tissue of origin, with implications for potential clinical application.
Figures
References
-
- Santos-Rosa H, Caldas C. Chromatin modifier enzymes, the histone code and cancer. Eur J Can. 2005. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

