Substrate specificity of bacterial oligosaccharyltransferase suggests a common transfer mechanism for the bacterial and eukaryotic systems
- PMID: 16641107
- PMCID: PMC1459022
- DOI: 10.1073/pnas.0509207103
Substrate specificity of bacterial oligosaccharyltransferase suggests a common transfer mechanism for the bacterial and eukaryotic systems
Abstract
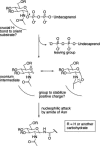
The PglB oligosaccharyltransferase (OTase) of Campylobacter jejuni can be functionally expressed in Escherichia coli, and its relaxed oligosaccharide substrate specificity allows the transfer of different glycans from the lipid carrier undecaprenyl pyrophosphate to an acceptor protein. To investigate the substrate specificity of PglB, we tested the transfer of a set of lipid-linked polysaccharides in E. coli and Salmonella enterica serovar Typhimurium. A hexose linked to the C-6 of the monosaccharide at the reducing end did not inhibit the transfer of the O antigen to the acceptor protein. However, PglB required an acetamido group at the C-2. A model for the mechanism of PglB involving this functional group was proposed. Previous experiments have shown that eukaryotic OTases have the same requirement, suggesting that eukaryotic and prokaryotic OTases catalyze the transfer of oligosaccharides by a conserved mechanism. Moreover, we demonstrated the functional transfer of the C. jejuni glycosylation system into S. enterica. The elucidation of the mechanism of action and the substrate specificity of PglB represents the foundation for engineering glycoproteins that will have an impact on biotechnology.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures

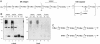

References
-
- Linton D., Allan E., Karlyshev A. V., Cronshaw A. D., Wren B. W. Mol. Microbiol. 2002;43:497–508. - PubMed
-
- Szymanski C. M., Yao R., Ewing C. P., Trust T. J., Guerry P. Mol. Microbiol. 1999;32:1022–1030. - PubMed
-
- Young N. M., Brisson J. R., Kelly J., Watson D. C., Tessier L., Lanthier P. H., Jarrell H. C., Cadotte N., St Michael F., Aberg E., Szymanski C. M. J. Biol. Chem. 2002;277:42530–42539. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

