NS1 interaction with CKII alpha: novel protein complex mediating parvovirus-induced cytotoxicity
- PMID: 16641266
- PMCID: PMC1472057
- DOI: 10.1128/JVI.80.10.4729-4739.2006
NS1 interaction with CKII alpha: novel protein complex mediating parvovirus-induced cytotoxicity
Abstract
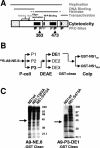
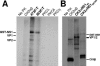
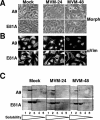
During a productive infection, the prototype strain of the parvovirus minute virus of mice (MVMp) induces dramatic morphological alterations in permissive A9 fibroblasts, culminating in cell lysis at the end of infection. These cytopathic effects (CPE) result from rearrangements and destruction of the cytoskeletal micro- and intermediate filaments, while other structures such as the nuclear lamina and particularly the microtubule network remain protected throughout the infection (J. P. F. Nüesch et al., Virology 331:159-174, 2005). In order to unravel the mechanism(s) by which parvoviruses trigger CPE, we searched for NS1 interaction partners by differential affinity chromatography, using distinct NS1 mutants debilitated specifically for this function. Thereby, we isolated an NS1 partner polypeptide, whose interaction with NS1 correlated with the competence of the viral product for CPE induction, and further identified it by tandem mass spectrometry and Western blotting analyses to consist of the catalytic subunit of casein kinase II, CKIIalpha. This interaction of NS1 with CKIIalpha suggested interference by the viral protein with intracellular signaling. Using permanent cell lines expressing dominant-negative CKIIalpha mutants, we were able to show that this kinase activity was indeed specifically involved in parvoviral CPE and progeny particle release. Furthermore, the NS1/CKIIalpha complex proved to be able to specifically phosphorylate viral capsids, indicating a mediator function of NS1 for CKII activity and specificity, at least in vitro. Altogether our data suggest that parvovirus-induced CPE is mediated by NS1 interference with intracellular CKII signaling.
Figures





References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

