Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir
- PMID: 16641282
- PMCID: PMC1472081
- DOI: 10.1128/JVI.80.10.4909-4920.2006
Emergence of CXCR4-using human immunodeficiency virus type 1 (HIV-1) variants in a minority of HIV-1-infected patients following treatment with the CCR5 antagonist maraviroc is from a pretreatment CXCR4-using virus reservoir
Abstract
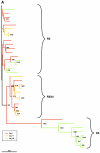
Antagonists of the human immunodeficiency virus type 1 (HIV-1) coreceptor, CCR5, are being developed as the first anti-HIV agents acting on a host cell target. We monitored the coreceptor tropism of circulating virus, screened at baseline for coreceptor tropism, in 64 HIV-1-infected patients who received maraviroc (MVC, UK-427,857) as monotherapy for 10 days. Sixty-two patients harbored CCR5-tropic virus at baseline and had a posttreatment phenotype result. Circulating virus remained CCR5 tropic in 60/62 patients, 51 of whom experienced an HIV RNA reduction from baseline of >1 log(10) copies/ml, indicating that CXCR4-using variants were not rapidly selected despite CCR5-specific drug pressure. In two patients, viral load declined during treatment and CXCR4-using virus was detected at day 11. No pretreatment factor predicted the emergence of CXCR4-tropic virus during maraviroc therapy in these two patients. Phylogenetic analysis of envelope (Env) clones from pre- and posttreatment time points indicated that the CXCR4-using variants probably emerged by outgrowth of a pretreatment CXCR4-using reservoir, rather than via coreceptor switch of a CCR5-tropic clone under selection pressure from maraviroc. Phylogenetic analysis was also performed on Env clones from a third patient harboring CXCR4-using virus prior to treatment. This patient was enrolled due to a sample labeling error. Although this patient experienced no overall reduction in viral load in response to treatment, the CCR5-tropic components of the circulating virus did appear to be suppressed while receiving maraviroc as monotherapy. Importantly, in all three patients, circulating virus reverted to predominantly CCR5 tropic following cessation of maraviroc.
Figures




References
-
- Berger, E. A., R. W. Doms, E. M. Fenyo, B. T. Korber, D. R. Littman, J. P. Moore, Q. J. Sattentau, H. Schuitemaker, J. Sodroski, and R. A. Weiss. 1998. A new classification for HIV-1. Nature 391:240. - PubMed
-
- Brumme, Z. L., W. W. Dong, B. Yip, B. Wynhoven, N. G. Hoffman, R. Swanstrom, M. A. Jensen, J. I. Mullins, R. S. Hogg, J. S. Montaner, and P. R. Harrigan. 2004. Clinical and immunological impact of HIV envelope V3 sequence variation after starting initial triple antiretroviral therapy. AIDS 18:F1-F9. - PubMed
-
- Brumme, Z. L., J. Goodrich, H. B. Mayer, C. J. Brumme, B. M. Henrick, B. Wynhoven, J. J. Asselin, P. K. Cheung, R. S. Hogg, J. S. Montaner, and P. R. Harrigan. 2005. Molecular and clinical epidemiology of CXCR4-using HIV-1 in a large population of antiretroviral-naive individuals. J. Infect. Dis. 192:466-474. - PubMed
-
- Coakley, E., C. J. Petropoulos, and J. M. Whitcomb. 2005. Assessing chemokine co-receptor usage in HIV. Curr. Opin. Infect. Dis. 18:9-15. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

