Predictive modelling of topology and loop variations in dimeric DNA quadruplex structures
- PMID: 16641317
- PMCID: PMC1449907
- DOI: 10.1093/nar/gkl182
Predictive modelling of topology and loop variations in dimeric DNA quadruplex structures
Abstract
We have used a combination of simulated annealing (SA), molecular dynamics (MD) and locally enhanced sampling (LES) methods in order to predict the favourable topologies and loop conformations of dimeric DNA quadruplexes with T2 or T3 loops. This follows on from our previous MD simulation studies on the influence of loop lengths on the topology of intramolecular quadruplex structures [P. Hazel et al. (2004) J. Am. Chem. Soc., 126, 16 405-16 415], which provided results consistent with biophysical data. The recent crystal structures of d(G4T3G4)2 and d(G4BrUT2G4) (P. Hazel et al. (2006) J. Am. Chem. Soc., in press) and the NMR-determined topology of d(TG4T2G4T)2 [A.T. Phan et al. (2004) J. Mol. Biol., 338, 93-102] have been used in the present study for comparison with simulation results. These together with MM-PBSA free-energy calculations indicate that lateral T3 loops are favoured over diagonal loops, in accordance with the experimental structures; however, distinct loop conformations have been predicted to be favoured compared to those found experimentally. Several lateral and diagonal loop conformations have been found to be similar in energy. The simulations suggest an explanation for the distinct patterns of observed dimer topology for sequences with T3 and T2 loops, which depend on the loop lengths, rather than only on G-quartet stability.
Figures


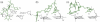
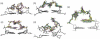
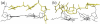

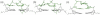

References
-
- Parkinson G.N., Lee M.P.H., Neidle S. Crystal structure of parallel quadruplexes from human telomeric DNA. Nature. 2002;417:876–880. - PubMed
-
- Ambrus A., Chen D., Dai J., Jones R.A., Yang D. Solution structure of the biologically relevant G-quadruplex element in the human c-myc promoter. Implications for G-quadruplex stabilization. Biochemistry. 2005;44:2048–2058. - PubMed

