Quantification of homozygosity in consanguineous individuals with autosomal recessive disease
- PMID: 16642444
- PMCID: PMC1474039
- DOI: 10.1086/503875
Quantification of homozygosity in consanguineous individuals with autosomal recessive disease
Abstract
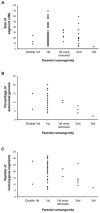
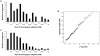
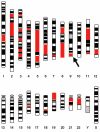
Individuals born of consanguineous union have segments of their genomes that are homozygous as a result of inheriting identical ancestral genomic segments through both parents. One consequence of this is an increased incidence of recessive disease within these sibships. Theoretical calculations predict that 6% (1/16) of the genome of a child of first cousins will be homozygous and that the average homozygous segment will be 20 cM in size. We assessed whether these predictions held true in populations that have preferred consanguineous marriage for many generations. We found that in individuals with a recessive disease whose parents were first cousins, on average, 11% of their genomes were homozygous (n = 38; range 5%-20%), with each individual bearing 20 homozygous segments exceeding 3 cM (n = 38; range of number of homozygous segments 7-32), and that the size of the homozygous segment associated with recessive disease was 26 cM (n = 100; range 5-70 cM). These data imply that prolonged parental inbreeding has led to a background level of homozygosity increased approximately 5% over and above that predicted by simple models of consanguinity. This has important clinical and research implications.
Figures
References
Web Resources
-
- Affymetrix, http://www.affymetrix.com/support/technical/datasheets/10k2_datasheet.pdf (for 10K SNP chip data)
-
- Center for Medical Genetics, http://research.marshfieldclinic.org/genetics/Map_Markers/maps/IndexMapF... (for Marshfield genetic maps for polymorphic microsatellite genetic locations)
-
- Geneservice, http://www.geneservice.co.uk/home/ (formerly MRC Geneservice, now an independent company)
-
- Online Mendelian Inheritance in Man (OMIM), http://www.ncbi.nlm.nih.gov/Omim/ (for Alstrom syndrome, Fuhrmann syndrome, Lebers amaurosis, nonsyndrome mental retardation, oro-facial-digital syndrome, retinitis pigmentosa, primary microcephaly, Seckel syndrome, and Warburg micro syndrome)
-
- UCSC Genome Bioinformatics, http://genome.ucsc.edu/ (for the Human Genome Browser and for physical order and location of markers and SNPs)
References
-
- Smith CAB (1974) Measures of homozygosity and inbreeding in populations. Ann Hum Genet 37:377–391 - PubMed
-
- Lander ES, Botstein D (1987) Homozygosity mapping: a way to map human recessive traits with the DNA of inbred children. Science 236:1567–1570 - PubMed
-
- Wright AF, Teague PW, Bruford E, Carothers A (1997) Problems in dealing with linkage heterogeneity in autosomal recessive forms of retinitis pigmentosa. In: Edwards JH, Pawlowitzki IH, Thompson E (eds) Genetic mapping of disease genes. Academic Press, London, pp 255–272
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources

