Flexible promoter architecture requirements for coactivator recruitment
- PMID: 16646957
- PMCID: PMC1488866
- DOI: 10.1186/1471-2199-7-16
Flexible promoter architecture requirements for coactivator recruitment
Abstract
Background: The spatial organization of transcription factor binding sites in regulatory DNA, and the composition of intersite sequences, influences the assembly of the multiprotein complexes that regulate RNA polymerase recruitment and thereby affects transcription. We have developed a genetic approach to investigate how reporter gene transcription is affected by varying the spacing between transcription factor binding sites. We characterized the components of promoter architecture that govern the yeast transcription factors Cbf1 and Met31/32, which bind independently, but collaboratively recruit the coactivator Met4.
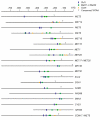
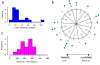
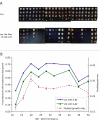
Results: A Cbf1 binding site was required upstream of a Met31/32 binding site for full reporter gene expression. Distance constraints on coactivator recruitment were more flexible than those for cooperatively binding transcription factors. Distances from 18 to 50 bp between binding sites support efficient recruitment of Met4, with only slight modulation by helical phasing. Intriguingly, we found that certain sequences located between the binding sites abolished gene expression.
Conclusion: These results yield insight to the influence of both binding site architecture and local DNA flexibility on gene expression, and can be used to refine computational predictions of gene expression from promoter sequences. In addition, our approach can be applied to survey promoter architecture requirements for arbitrary combinations of transcription factor binding sites.
Figures


References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

