Eight previously unidentified mutations found in the OA1 ocular albinism gene
- PMID: 16646960
- PMCID: PMC1468396
- DOI: 10.1186/1471-2350-7-41
Eight previously unidentified mutations found in the OA1 ocular albinism gene
Abstract
Background: Ocular albinism type 1 (OA1) is an X-linked ocular disorder characterized by a severe reduction in visual acuity, nystagmus, hypopigmentation of the retinal pigmented epithelium, foveal hypoplasia, macromelanosomes in pigmented skin and eye cells, and misrouting of the optical tracts. This disease is primarily caused by mutations in the OA1 gene.
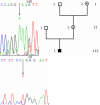
Methods: The ophthalmologic phenotype of the patients and their family members was characterized. We screened for mutations in the OA1 gene by direct sequencing of the nine PCR-amplified exons, and for genomic deletions by PCR-amplification of large DNA fragments.
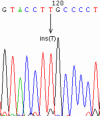
Results: We sequenced the nine exons of the OA1 gene in 72 individuals and found ten different mutations in seven unrelated families and three sporadic cases. The ten mutations include an amino acid substitution and a premature stop codon previously reported by our team, and eight previously unidentified mutations: three amino acid substitutions, a duplication, a deletion, an insertion and two splice-site mutations. The use of a novel Taq polymerase enabled us to amplify large genomic fragments covering the OA1 gene. and to detect very likely six distinct large deletions. Furthermore, we were able to confirm that there was no deletion in twenty one patients where no mutation had been found.
Conclusion: The identified mutations affect highly conserved amino acids, cause frameshifts or alternative splicing, thus affecting folding of the OA1 G protein coupled receptor, interactions of OA1 with its G protein and/or binding with its ligand.
Figures

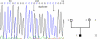
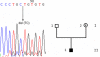
Similar articles
-
Mutational analysis of the OA1 gene in ocular albinism.Ophthalmic Genet. 2003 Sep;24(3):167-73. doi: 10.1076/opge.24.3.167.15605. Ophthalmic Genet. 2003. PMID: 12868035
-
New insights into ocular albinism type 1 (OA1): Mutations and polymorphisms of the OA1 gene.Hum Mutat. 2002 Feb;19(2):85-92. doi: 10.1002/humu.10034. Hum Mutat. 2002. PMID: 11793467 Review.
-
Diverse prevalence of large deletions within the OA1 gene in ocular albinism type 1 patients from Europe and North America.Hum Genet. 2001 Jan;108(1):51-4. doi: 10.1007/s004390000440. Hum Genet. 2001. PMID: 11214907
-
Analysis of the OA1 gene reveals mutations in only one-third of patients with X-linked ocular albinism.Hum Mol Genet. 1995 Dec;4(12):2319-25. doi: 10.1093/hmg/4.12.2319. Hum Mol Genet. 1995. PMID: 8634705
-
Ocular albinism type 1: more than meets the eye.Pigment Cell Res. 2001 Aug;14(4):243-8. doi: 10.1034/j.1600-0749.2001.140403.x. Pigment Cell Res. 2001. PMID: 11549106 Review.
Cited by
-
CRISPR-AsCas12a Efficiently Corrects a GPR143 Intronic Mutation in Induced Pluripotent Stem Cells from an Ocular Albinism Patient.CRISPR J. 2022 Jun;5(3):457-471. doi: 10.1089/crispr.2021.0110. CRISPR J. 2022. PMID: 35686978 Free PMC article.
-
Clinical utility gene card for oculocutaneous (OCA) and ocular albinism (OA)-an update.Eur J Hum Genet. 2021 Oct;29(10):1577-1583. doi: 10.1038/s41431-021-00809-w. Epub 2021 Jan 27. Eur J Hum Genet. 2021. PMID: 33504991 Free PMC article. No abstract available.
-
Identification of a novel GPR143 mutation in a large Chinese family with congenital nystagmus as the most prominent and consistent manifestation.J Hum Genet. 2007;52(6):565-570. doi: 10.1007/s10038-007-0152-3. Epub 2007 May 22. J Hum Genet. 2007. PMID: 17516023
-
Novel GPR143 mutations and clinical characteristics in six Chinese families with X-linked ocular albinism.Mol Vis. 2008;14:1974-82. Epub 2008 Oct 30. Mol Vis. 2008. PMID: 18978956 Free PMC article.
-
A novel nonsense mutation of the GPR143 gene identified in a Chinese pedigree with ocular albinism.PLoS One. 2012;7(8):e43177. doi: 10.1371/journal.pone.0043177. Epub 2012 Aug 20. PLoS One. 2012. PMID: 22916221 Free PMC article.
References
-
- O'Donnell FE, Jr, Hambrick GW, Jr, Green WR, Iliff WJ, Stone DL. X-linked ocular albinism. An oculocutaneous macromelanosomal disorder. Arch Ophthalmol. 1976;94:1883–1892. - PubMed
-
- Garner A, Jay BS. Macromelanosomes in X-linked ocular albinism. Histopathology. 1980;4:243–254. - PubMed
-
- Cortese K, Giordano F, Surace EM, Venturi C, Ballabio A, Tacchetti C, Marigo V. The Ocular Albinism Type 1 (OA1) Gene Controls Melanosome Maturation and Size. IOVS. 2005;46:4358–4364. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

