Metabolomic database annotations via query of elemental compositions: mass accuracy is insufficient even at less than 1 ppm
- PMID: 16646969
- PMCID: PMC1464138
- DOI: 10.1186/1471-2105-7-234
Metabolomic database annotations via query of elemental compositions: mass accuracy is insufficient even at less than 1 ppm
Abstract
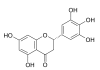
Background: Metabolomic studies are targeted at identifying and quantifying all metabolites in a given biological context. Among the tools used for metabolomic research, mass spectrometry is one of the most powerful tools. However, metabolomics by mass spectrometry always reveals a high number of unknown compounds which complicate in depth mechanistic or biochemical understanding. In principle, mass spectrometry can be utilized within strategies of de novo structure elucidation of small molecules, starting with the computation of the elemental composition of an unknown metabolite using accurate masses with errors <5 ppm (parts per million). However even with very high mass accuracy (<1 ppm) many chemically possible formulae are obtained in higher mass regions. In automatic routines an additional orthogonal filter therefore needs to be applied in order to reduce the number of potential elemental compositions. This report demonstrates the necessity of isotope abundance information by mathematical confirmation of the concept.
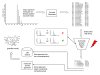
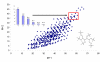
Results: High mass accuracy (<1 ppm) alone is not enough to exclude enough candidates with complex elemental compositions (C, H, N, S, O, P, and potentially F, Cl, Br and Si). Use of isotopic abundance patterns as a single further constraint removes >95% of false candidates. This orthogonal filter can condense several thousand candidates down to only a small number of molecular formulas. Example calculations for 10, 5, 3, 1 and 0.1 ppm mass accuracy are given. Corresponding software scripts can be downloaded from http://fiehnlab.ucdavis.edu. A comparison of eight chemical databases revealed that PubChem and the Dictionary of Natural Products can be recommended for automatic queries using molecular formulae.
Conclusion: More than 1.6 million molecular formulae in the range 0-500 Da were generated in an exhaustive manner under strict observation of mathematical and chemical rules. Assuming that ion species are fully resolved (either by chromatography or by high resolution mass spectrometry), we conclude that a mass spectrometer capable of 3 ppm mass accuracy and 2% error for isotopic abundance patterns outperforms mass spectrometers with less than 1 ppm mass accuracy or even hypothetical mass spectrometers with 0.1 ppm mass accuracy that do not include isotope information in the calculation of molecular formulae.
Figures


References
-
- Weckwerth W, Wenzel K, Fiehn O. Process for the integrated extraction, identification and quantification of metabolites, proteins and RNA to reveal their co-regulation in biochemical networks. Proteomics. 2004;4:78–83. - PubMed
-
- Nicolaou KC, Snyder , Scott A. Chasing molecules that were never there: Misassigned natural products and the role of chemical synthesis in modern structure elucidation. Angew Chem Int Ed. 2005;44:1012–1044. - PubMed
-
- Wagner C, Sefkow M, Kopka J. Construction and application of a mass spectral and retention time index database generated from plant GC/EI-TOF-MS metabolite profiles. Phytochemistry. 2003;62:887–900. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

