Chlororespiratory reduction 6 is a novel factor required for accumulation of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis
- PMID: 16648216
- PMCID: PMC1475432
- DOI: 10.1104/pp.106.080267
Chlororespiratory reduction 6 is a novel factor required for accumulation of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis
Abstract
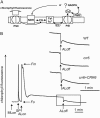
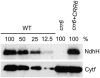
The chloroplast NAD(P)H dehydrogenase (NDH) complex is involved in photosystem I cyclic electron transport and chlororespiration in higher plants. An Arabidopsis (Arabidopsis thaliana) chlororespiratory reduction 6 (crr6) mutant lacking NDH activity was identified by means of chlorophyll fluorescence imaging. Accumulation of the NDH complex was impaired in crr6. Physiological characterization of photosynthetic electron transport indicated the specific defect of the NDH complex in crr6. In contrast to the CRR7 protein that was recently identified as a potential novel subunit of the NDH complex by means of the same screening, the CRR6 protein was stable under the crr2 mutant background in which the NDH complex does not accumulate. The CRR6 gene (At2g47910) encodes a novel protein without any known motif. Although CRR6 does not have any transmembrane domains, it is localized in the thylakoid membrane fraction of the chloroplast. CRR6 is conserved in phototrophs, including cyanobacteria, from which the chloroplast NDH complex has evolutionally originated, but not in Chlamydomonas reinhardtii, in which the NDH complex is absent. We believe that CRR6 is a novel specific factor for the assembly or stabilization of the NDH complex.
Figures


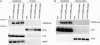

Similar articles
-
Identification of a novel protein, CRR7, required for the stabilization of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis.Plant J. 2005 Dec;44(6):1036-44. doi: 10.1111/j.1365-313X.2005.02604.x. Plant J. 2005. PMID: 16359395
-
Chloroplast stromal proteins, CRR6 and CRR7, are required for assembly of the NAD(P)H dehydrogenase subcomplex A in Arabidopsis.Plant J. 2010 Jul;63(2):203-211. doi: 10.1111/j.1365-313X.2010.04240.x. Epub 2010 Apr 28. Plant J. 2010. Retraction in: Plant J. 2023 Apr;114(2):455. doi: 10.1111/tpj.16170. PMID: 20444231 Retracted.
-
A eukaryotic factor required for accumulation of the chloroplast NAD(P)H dehydrogenase complex in Arabidopsis.Plant Physiol. 2006 Dec;142(4):1683-9. doi: 10.1104/pp.106.088682. Epub 2006 Oct 13. Plant Physiol. 2006. PMID: 17041026 Free PMC article.
-
Towards characterization of the chloroplast NAD(P)H dehydrogenase complex.Mol Plant. 2009 Nov;2(6):1127-40. doi: 10.1093/mp/ssp052. Epub 2009 Jul 21. Mol Plant. 2009. PMID: 19995722 Review.
-
Chloroplast NDH: A different enzyme with a structure similar to that of respiratory NADH dehydrogenase.Biochim Biophys Acta. 2016 Jul;1857(7):1015-22. doi: 10.1016/j.bbabio.2015.10.013. Epub 2015 Oct 28. Biochim Biophys Acta. 2016. PMID: 26519774 Review.
Cited by
-
Chlororespiration and grana hyperstacking: how an Arabidopsis double mutant can survive despite defects in starch biosynthesis and daily carbon export from chloroplasts.Plant Physiol. 2009 Jan;149(1):515-33. doi: 10.1104/pp.108.128124. Epub 2008 Oct 31. Plant Physiol. 2009. PMID: 18978072 Free PMC article.
-
Quantitative analysis of the relationship between induction kinetics of chlorophyll fluorescence and function of genes in the cyanobacterium Synechocystis sp. PCC 6803.Photosynth Res. 2009 Jul;101(1):47-58. doi: 10.1007/s11120-009-9462-y. Epub 2009 Jul 1. Photosynth Res. 2009. PMID: 19568952
-
Structure, function, and evolution of the PsbP protein family in higher plants.Photosynth Res. 2008 Oct-Dec;98(1-3):427-37. doi: 10.1007/s11120-008-9359-1. Epub 2008 Sep 13. Photosynth Res. 2008. PMID: 18791807 Review.
-
A model for describing the light response of the nonphotochemical quenching of chlorophyll fluorescence.Photosynth Res. 2011 May;108(1):61-76. doi: 10.1007/s11120-011-9654-0. Epub 2011 Apr 23. Photosynth Res. 2011. PMID: 21516348
-
Distinct functions for the two PsbP-like proteins PPL1 and PPL2 in the chloroplast thylakoid lumen of Arabidopsis.Plant Physiol. 2007 Nov;145(3):668-79. doi: 10.1104/pp.107.105866. Epub 2007 Sep 7. Plant Physiol. 2007. PMID: 17827269 Free PMC article.
References
-
- Barkan A, Goldschmidt-Clermont M (2000) Participation of nuclear genes in chloroplast gene expression. Biochimie 82: 559–572 - PubMed
-
- Battchikova N, Zhang P, Rudd S, Ogawa T, Aro E-M (2005) Identification of NdhL and Ssl1690 (NdhO) in NDH-1L and NDH-1M complexes of Synechocystis sp. PCC 6803. J Biol Chem 280: 2587–2595 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

