Diffusion of isolated DNA molecules: dependence on length and topology
- PMID: 16648255
- PMCID: PMC1450111
- DOI: 10.1073/pnas.0601903103
Diffusion of isolated DNA molecules: dependence on length and topology
Abstract
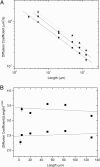
The conformation and dynamics of circular polymers is a subject of considerable theoretical and experimental interest. DNA is an important example because it occurs naturally in different topological states, including linear, relaxed circular, and supercoiled circular forms. A fundamental question is how the diffusion coefficients of isolated polymers scale with molecular length and how they vary for different topologies. Here, diffusion coefficients D for relaxed circular, supercoiled, and linear DNA molecules of length L ranging from approximately 6 to 290 kbp were measured by tracking the Brownian motion of single molecules. A topology-independent scaling law D approximately L(-nu) was observed with nu(L) = 0.571 +/- 0.014, nu(C) = 0.589 +/- 0.018, and nu(S) = 0.571 +/- 0.057 for linear, relaxed circular, and supercoiled DNA, respectively, in good agreement with the scaling exponent of nu congruent with 0.588 predicted by renormalization group theory for polymers with significant excluded volume interactions. Our findings thus provide evidence in support of several theories that predict an effective diameter of DNA much greater than the Debye screening length. In addition, the measured ratio D(Circular)/D(Linear) = 1.32 +/- 0.014 was closer to the value of 1.45 predicted by using renormalization group theory than the value of 1.18 predicted by classical Kirkwood hydrodynamic theory and agreed well with a value of 1.31 predicted when incorporating a recently proposed expression for the radius of gyration of circular polymers into the Zimm model.
Conflict of interest statement
Conflict of interest statement: No conflicts declared.
Figures
References
-
- Alberts B., Johnson A., Lewis J., Raff M., Roberts K., Walter P. Molecular Biology of the Cell. New York: Garland; 2002.
-
- Doi M., Edwards S. The Theory of Polymer Dynamics. Oxford: Clarendon; 1986.
-
- Jagodzinski O., Eisenriegler E., Kremer K. J. Phys. I [French] 1992;2:2243–2279.
-
- Koniaris K., Muthukumar M. J. Chem. Phys. 1991;95:2873–2881.
-
- Shimamura M. K., Deguchi T. Phys. Rev. E. 2001;64:020801. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

