A sequence motif conserved in diverse nuclear proteins identifies a protein interaction domain utilised for nuclear targeting by human TFIIS
- PMID: 16648364
- PMCID: PMC1450333
- DOI: 10.1093/nar/gkl239
A sequence motif conserved in diverse nuclear proteins identifies a protein interaction domain utilised for nuclear targeting by human TFIIS
Abstract
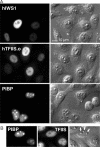
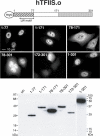
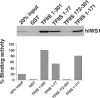
The three structural domains of transcription elongation factor TFIIS are conserved from yeast to human. Although the N-terminal domain is not needed for transcriptional activity, a similar sequence has been identified previously in other transcription factors. We found this conserved sequence, the LW motif, in another three human proteins that are predominantly nuclear localized. We investigated two examples to determine whether the LW motif is actually a dedicated nuclear targeting signal. However, in one of the newly identified proteins, hIWS1 (human Iws1), a region containing classic nuclear localization signals (NLS) rather than the LW motif was necessary and sufficient for nuclear targeting in HeLa cells. In contrast, human TFIIS does not possess an NLS and only constructs containing the LW motif were efficiently targeted to nuclei. Moreover, mutations in the motif could cause cytoplasmic accumulation of TFIIS and enabled a structure/function assay for the domain based on the efficiency of nuclear targeting. Finally, GST pull-down assays showed that the LW motif is part of a protein-binding domain. We suggest that the targeting role the LW motif plays in TFIIS arises from its more general function as a protein interaction domain, enabling TFIIS to bind a carrier protein(s) that accomplishes nuclear import.
Figures



Similar articles
-
The structure of an Iws1/Spt6 complex reveals an interaction domain conserved in TFIIS, Elongin A and Med26.EMBO J. 2010 Dec 1;29(23):3979-91. doi: 10.1038/emboj.2010.272. Epub 2010 Nov 5. EMBO J. 2010. PMID: 21057455 Free PMC article.
-
Structural basis for evolutionarily conserved interactions between TFIIS and Paf1C.Int J Biol Macromol. 2023 Dec 31;253(Pt 2):126764. doi: 10.1016/j.ijbiomac.2023.126764. Epub 2023 Sep 9. Int J Biol Macromol. 2023. PMID: 37696373 Free PMC article.
-
An extended bipartite nuclear localization signal in Smad4 is required for its nuclear import and transcriptional activity.Oncogene. 2003 Feb 20;22(7):1057-69. doi: 10.1038/sj.onc.1206212. Oncogene. 2003. PMID: 12592392
-
The TFIIS N-terminal domain (TND): a transcription assembly module at the interface of order and disorder.Biochem Soc Trans. 2023 Feb 27;51(1):125-135. doi: 10.1042/BST20220342. Biochem Soc Trans. 2023. PMID: 36651856 Free PMC article. Review.
-
Domain acquisition enabled functional expansion of the TFIIS transcription factor family.Cell Biosci. 2025 Jun 4;15(1):78. doi: 10.1186/s13578-025-01423-9. Cell Biosci. 2025. PMID: 40468406 Free PMC article. Review.
Cited by
-
The structure of an Iws1/Spt6 complex reveals an interaction domain conserved in TFIIS, Elongin A and Med26.EMBO J. 2010 Dec 1;29(23):3979-91. doi: 10.1038/emboj.2010.272. Epub 2010 Nov 5. EMBO J. 2010. PMID: 21057455 Free PMC article.
-
Crystallization and preliminary crystallographic analysis of eukaryotic transcription and mRNA export factor Iws1 from Encephalitozoon cuniculi.Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010 Feb 1;66(Pt 2):207-10. doi: 10.1107/S1744309109052749. Epub 2010 Jan 28. Acta Crystallogr Sect F Struct Biol Cryst Commun. 2010. PMID: 20124725 Free PMC article.
-
Arabidopsis IWS1 interacts with transcription factor BES1 and is involved in plant steroid hormone brassinosteroid regulated gene expression.Proc Natl Acad Sci U S A. 2010 Feb 23;107(8):3918-23. doi: 10.1073/pnas.0909198107. Epub 2010 Feb 5. Proc Natl Acad Sci U S A. 2010. PMID: 20139304 Free PMC article.
-
Structural basis for evolutionarily conserved interactions between TFIIS and Paf1C.Int J Biol Macromol. 2023 Dec 31;253(Pt 2):126764. doi: 10.1016/j.ijbiomac.2023.126764. Epub 2023 Sep 9. Int J Biol Macromol. 2023. PMID: 37696373 Free PMC article.
-
Identification and characterization of two trypanosome TFIIS proteins exhibiting particular domain architectures and differential nuclear localizations.Mol Microbiol. 2008 Sep;69(5):1121-36. doi: 10.1111/j.1365-2958.2008.06348.x. Epub 2008 Jul 4. Mol Microbiol. 2008. PMID: 18627464 Free PMC article.
References
-
- Proudfoot N. Connecting transcription to messenger RNA processing. Trends Biochem. Sci. 2000;25:290–293. - PubMed
-
- Neugebauer K.M. On the importance of being co-transcriptional. J. Cell Sci. 2002;115:3865–3871. - PubMed
-
- Maniatis T., Reed R. An extensive network of coupling among gene expression machines. Nature. 2002;416:499–506. - PubMed
-
- Lewis J.D., Tollervey D. Like attracts like: getting RNA processing together in the nucleus. Science. 2000;288:1385–1389. - PubMed
-
- Lamond A.I., Spector D.L. Nuclear speckles: a model for nuclear organelles. Nature Rev. Mol. Cell Biol. 2003;4:605–612. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

